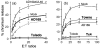Modulation of natural killer cells by human cytomegalovirus - PubMed (original) (raw)
Review
doi: 10.1016/j.jcv.2007.10.027.
Peter Tomasec, Richard J Stanton, Melanie Armstrong, Virginie Prod'homme, Rebecca Aicheler, Brian P McSharry, Carole R Rickards, Daniel Cochrane, Sian Llewellyn-Lacey, Eddie C Y Wang, Cora A Griffin, Andrew J Davison
Affiliations
- PMID: 18069056
- PMCID: PMC2843162
- DOI: 10.1016/j.jcv.2007.10.027
Review
Modulation of natural killer cells by human cytomegalovirus
Gavin W G Wilkinson et al. J Clin Virol. 2008 Mar.
Abstract
Human cytomegalovirus (HCMV) causes lifelong, persistent infections and its survival is under intense, continuous selective pressure from the immune system. A key aspect of HCMV's capacity for survival lies in immune avoidance. In this context, cells undergoing productive infection exhibit remarkable resistance to natural killer (NK) cell-mediated cytolysis in vitro. To date, six genes encoding proteins (UL16, UL18, UL40, UL83, UL141 and UL142) and one encoding a microRNA (miR-UL112) have been identified as capable of suppressing NK cell recognition. Even though HCMV infection efficiently activates expression of ligands for the NK cell activating receptor NKG2D, at least three functions (UL16, UL142 and miR-UL112) act in concert to suppress presentation of these ligands on the cell surface. Although HCMV downregulates expression of endogenous MHC-I, it encodes an MHC-I homologue (UL18) and also upregulates the expression of cellular HLA-E through the action of UL40. The disruption of normal intercellular connections exposes ligands for NK cell activating receptors on the cell surface, notably CD155. HCMV overcomes this vulnerability by encoding a function (UL141) that acts post-translationally to suppress cell surface expression of CD155. The mechanisms by which HCMV systematically evades (or, more properly, modulates) NK cell recognition constitutes an area of growing understanding that is enhancing our appreciation of the basic mechanisms of NK cell function in humans.
Figures
Fig. 1
Infection with low passage HCMV strains provides maximum protection against NK cell attack. (a) HFs infected with high passage strain AD169 show significant protection from NK cell lysis, compared with uninfected targets or targets infected with an AD169 UL40 deletion mutant. Cells infected with strain Toledo or other low passage isolates (not shown) were completely resistant to NK cell attack. (b) The ability to induce complete resistance to NK cell lysis could be restored by inserting the strain Toledo UL_b_′ sequence into high passage strain Towne to produce recombinant Tx4 (kindly provided by E. Mocarski). Modified from Tomasec et al. (2005).
Fig. 2
Upregulation of HLA-E by HCMV gpUL40. HLA-E exhibits only minor allelic variation and binds a conserved nonameric peptide derived from the leader sequence of HLA-A, -B, -C and -G in a TAP-dependent manner. The HCMV US6 protein binds TAP to inhibit peptide transport. Consequently, US6 inhibits both MHC-I antigen presentation and surface expression of HLA-E. However, the leader sequence of gpUL40 also encodes an HLA-E-binding peptide that is released to the ER independently of TAP. UL40 thus upregulates cell surface expression of HLA-E, a recognized ligand for the NK cell inhibitory receptor CD94/NKG2A.
Fig. 3
Gene map of the HCMV strain Merlin genome. Inverted repeat regions are shown in a thicker format than the two unique regions. Protein-coding regions are indicated by coloured arrows grouped according to the key, with gene nomenclature below. Introns are shown as narrow white bars. Genes whose names commence with RL, TRS and IRS are given in full, but the UL and US prefixes have been omitted from UL1-UL150 (12–194 kbp) and US1-US34A (199–231 kbp). Colours differentiate between genes on the basis of conservation among the Alpha, _Beta_- and Gammaherpesvirinae (core genes) or between the _Beta_- and Gammaherpesvirinae (sub-core genes), with subsets of the remaining non-core genes grouped into gene families. The location of the UL/_b_′ region (UL148 to UL150) is highlighted by thick horizontal lines. Modified from Dolan et al. (2004) by permission of the Society for General Microbiology.
Similar articles
- Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein.
Rölle A, Mousavi-Jazi M, Eriksson M, Odeberg J, Söderberg-Nauclér C, Cosman D, Kärre K, Cerboni C. Rölle A, et al. J Immunol. 2003 Jul 15;171(2):902-8. doi: 10.4049/jimmunol.171.2.902. J Immunol. 2003. PMID: 12847260 - Genetic Variability of Human Cytomegalovirus Clinical Isolates Correlates With Altered Expression of Natural Killer Cell-Activating Ligands and IFN-γ.
Galitska G, Coscia A, Forni D, Steinbrueck L, De Meo S, Biolatti M, De Andrea M, Cagliani R, Leone A, Bertino E, Schulz T, Santoni A, Landolfo S, Sironi M, Cerboni C, Dell'Oste V. Galitska G, et al. Front Immunol. 2021 Apr 9;12:532484. doi: 10.3389/fimmu.2021.532484. eCollection 2021. Front Immunol. 2021. PMID: 33897679 Free PMC article. - Expression of the UL16 glycoprotein of Human Cytomegalovirus protects the virus-infected cell from attack by natural killer cells.
Valés-Gómez M, Browne H, Reyburn HT. Valés-Gómez M, et al. BMC Immunol. 2003 Mar 14;4:4. doi: 10.1186/1471-2172-4-4. Epub 2003 Mar 14. BMC Immunol. 2003. PMID: 12694635 Free PMC article. - HCMV-Encoded NK Modulators: Lessons From in vitro and in vivo Genetic Variation.
Patel M, Vlahava VM, Forbes SK, Fielding CA, Stanton RJ, Wang ECY. Patel M, et al. Front Immunol. 2018 Oct 1;9:2214. doi: 10.3389/fimmu.2018.02214. eCollection 2018. Front Immunol. 2018. PMID: 30327650 Free PMC article. Review. - The UL16-binding proteins, a novel family of MHC class I-related ligands for NKG2D, activate natural killer cell functions.
Sutherland CL, Chalupny NJ, Cosman D. Sutherland CL, et al. Immunol Rev. 2001 Jun;181:185-92. doi: 10.1034/j.1600-065x.2001.1810115.x. Immunol Rev. 2001. PMID: 11513139 Review.
Cited by
- KIR and HLA under pressure: evidences of coevolution across worldwide populations.
Augusto DG, Petzl-Erler ML. Augusto DG, et al. Hum Genet. 2015 Sep;134(9):929-40. doi: 10.1007/s00439-015-1579-9. Epub 2015 Jun 23. Hum Genet. 2015. PMID: 26099314 Review. - The human cytomegalovirus protein UL147A downregulates the most prevalent MICA allele: MICA*008, to evade NK cell-mediated killing.
Seidel E, Dassa L, Schuler C, Oiknine-Djian E, Wolf DG, Le-Trilling VTK, Mandelboim O. Seidel E, et al. PLoS Pathog. 2021 May 3;17(5):e1008807. doi: 10.1371/journal.ppat.1008807. eCollection 2021 May. PLoS Pathog. 2021. PMID: 33939764 Free PMC article. - Diverse immune evasion strategies by human cytomegalovirus.
Noriega V, Redmann V, Gardner T, Tortorella D. Noriega V, et al. Immunol Res. 2012 Dec;54(1-3):140-51. doi: 10.1007/s12026-012-8304-8. Immunol Res. 2012. PMID: 22454101 Review. - The latency-associated UL138 gene product of human cytomegalovirus sensitizes cells to tumor necrosis factor alpha (TNF-alpha) signaling by upregulating TNF-alpha receptor 1 cell surface expression.
Montag C, Wagner JA, Gruska I, Vetter B, Wiebusch L, Hagemeier C. Montag C, et al. J Virol. 2011 Nov;85(21):11409-21. doi: 10.1128/JVI.05028-11. Epub 2011 Aug 31. J Virol. 2011. PMID: 21880774 Free PMC article. - Multiparametric analysis of host response to murine cytomegalovirus in MHC class I-disparate mice reveals primacy of Dk-licensed Ly49G2+ NK cells in viral control.
Prince J, Lundgren A, Stadnisky MD, Nash WT, Beeber A, Turner SD, Brown MG. Prince J, et al. J Immunol. 2013 Nov 1;191(9):4709-19. doi: 10.4049/jimmunol.1301388. Epub 2013 Sep 25. J Immunol. 2013. PMID: 24068668 Free PMC article.
References
- Arnon TI, Achdout H, Levi O, Markel G, Saleh N, Katz G, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–23. - PubMed
- Beck S, Barrell BG. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature. 1988;331:269–72. - PubMed
- Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–5. - PubMed
- Browne H, Smith G, Beck S, Minson T. A complex between the MHC class I homologue encoded by human cytomegalovirus and beta 2 microglobulin. Nature. 1990;347:770–2. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0300180/MRC_/Medical Research Council/United Kingdom
- G0700142/MRC_/Medical Research Council/United Kingdom
- G0500617(74644)/MRC_/Medical Research Council/United Kingdom
- G0300180(65735)/MRC_/Medical Research Council/United Kingdom
- MC_UU_12014/3/MRC_/Medical Research Council/United Kingdom
- G0500617/MRC_/Medical Research Council/United Kingdom
- MC_U130184136/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials