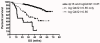Genetic events in the pathogenesis of multiple myeloma - PubMed (original) (raw)
Review
Genetic events in the pathogenesis of multiple myeloma
W J Chng et al. Best Pract Res Clin Haematol. 2007 Dec.
Abstract
The genetics of myeloma has been increasingly elucidated in recent years. Recurrent genetic events, and also biologically distinct and clinically relevant genetic subtypes of myeloma have been defined. This has facilitated our understanding of the molecular pathogenesis of the disease. In addition, some genetic abnormalities have proved to be highly reproducible prognostic factors. With the expanding therapeutic armamentarium, it is time to include genetic assessment as part of clinical evaluation of myeloma patients to guide management. In this review we examine the role of various genetic abnormalities in the molecular pathogenesis of myeloma, and the use of such abnormalities in disease classification, prognosis and clinical management.
Figures
Figure 1
Disease stages and timing of oncogenic events (see text for further details). The degree of intersection between the overlapping triangles estimates the percentage of each genetic subgroup with coexisting genetic abnormalities. The translocation partners in the IgH translocation (TLC) group are ordered according to increasing frequency of concurrent Δ13. The different mutations activating the different signaling or cell-cycle regulatory pathways are mutually exclusive, i.e., RAS and FGFR3 mutations always occur in different patients. *The 8q24 partner referred to here is MAFA; C-MYC is also located on 8q24, but these are usually secondary IgH TLCs.
Figure 2
The t(4;14) translocation further dissects the survival of high-risk patients defined by 17-gene high-risk molecular signature. Combination of t(4;14) and high risk defined by the 17-gene model (logQ4/Q1 > 0.85) identifies three groups of patients with significantly different survival (log-rank P < 0.0001) in patients entered into total therapy II (n = 351): a high-risk group (n = 16), defined by presence of logQ4/Q1 > 0.85 and t(4;14), with a median overall survival of 16.7 months; an intermediate-risk group (n = 45), defined by the presence of logQ4/Q1 > 0.85 without t(4;14), with a median overall survival of 39.8 months; and a low-risk group (n = 289), defined by logQ4/Q1 < 0.85, with a median survival not yet reached. (Gene expression and survival data available from the NCBI Gene Expression Omnibus [GEO] using accession number GSE2658.) For patients with logQ4/Q1 < 0.85, the presence of t(4;14) did not significantly alter survival and is therefore combined for survival analysis.
Similar articles
- [Study of the etiologic pathogenesis of multiple myeloma].
Hou J, Kong X. Hou J, et al. Zhonghua Xue Ye Xue Za Zhi. 1998 Nov;19(11):607-9. Zhonghua Xue Ye Xue Za Zhi. 1998. PMID: 11263345 Review. Chinese. No abstract available. - Role of genetics in prognostication in myeloma.
Avet-Loiseau H. Avet-Loiseau H. Best Pract Res Clin Haematol. 2007 Dec;20(4):625-35. doi: 10.1016/j.beha.2007.08.005. Best Pract Res Clin Haematol. 2007. PMID: 18070710 Review. - [Subclassification of multiple myeloma based on the partner genes of immunoglobulin heavy chain gene translocations].
Nishida K, Taniwaki M. Nishida K, et al. Rinsho Ketsueki. 2000 May;41(5):414-7. Rinsho Ketsueki. 2000. PMID: 10879102 Review. Japanese. No abstract available. - Heavy/light chain ratio normalization prior to transplant is of independent prognostic significance in multiple myeloma: a BMT CTN 0102 correlative study.
D'Souza A, Pasquini M, Logan B, Giralt S, Krishnan A, Antin J, Howard A, Goodman S, Qazilbash M, Knust K, Sahebi F, Weisdorf D, Vesole D, Stadtmauer E, Maloney D, Hari P. D'Souza A, et al. Br J Haematol. 2017 Sep;178(5):816-819. doi: 10.1111/bjh.14170. Epub 2016 Jun 13. Br J Haematol. 2017. PMID: 27292583 Free PMC article. No abstract available. - MGUS and smoldering multiple myeloma: update on pathogenesis, natural history, and management.
Rajkumar SV. Rajkumar SV. Hematology Am Soc Hematol Educ Program. 2005:340-5. doi: 10.1182/asheducation-2005.1.340. Hematology Am Soc Hematol Educ Program. 2005. PMID: 16304401
Cited by
- The PI3K/Akt signaling pathway regulates the expression of Hsp70, which critically contributes to Hsp90-chaperone function and tumor cell survival in multiple myeloma.
Chatterjee M, Andrulis M, Stühmer T, Müller E, Hofmann C, Steinbrunn T, Heimberger T, Schraud H, Kressmann S, Einsele H, Bargou RC. Chatterjee M, et al. Haematologica. 2013 Jul;98(7):1132-41. doi: 10.3324/haematol.2012.066175. Epub 2012 Oct 12. Haematologica. 2013. PMID: 23065523 Free PMC article. - Identification of unbalanced genome copy number abnormalities in patients with multiple myeloma by single-nucleotide polymorphism genotyping microarray analysis.
Kamada Y, Sakata-Yanagimoto M, Sanada M, Sato-Otsubo A, Enami T, Suzukawa K, Kurita N, Nishikii H, Yokoyama Y, Okoshi Y, Hasegawa Y, Ogawa S, Chiba S. Kamada Y, et al. Int J Hematol. 2012 Oct;96(4):492-500. doi: 10.1007/s12185-012-1171-1. Epub 2012 Sep 13. Int J Hematol. 2012. PMID: 22972171 - Transcriptional repression by the HDAC4-RelB-p52 complex regulates multiple myeloma survival and growth.
Vallabhapurapu SD, Noothi SK, Pullum DA, Lawrie CH, Pallapati R, Potluri V, Kuntzen C, Khan S, Plas DR, Orlowski RZ, Chesi M, Kuehl WM, Bergsagel PL, Karin M, Vallabhapurapu S. Vallabhapurapu SD, et al. Nat Commun. 2015 Oct 12;6:8428. doi: 10.1038/ncomms9428. Nat Commun. 2015. PMID: 26455434 - The t(14;20) is a poor prognostic factor in myeloma but is associated with long-term stable disease in monoclonal gammopathies of undetermined significance.
Ross FM, Chiecchio L, Dagrada G, Protheroe RK, Stockley DM, Harrison CJ, Cross NC, Szubert AJ, Drayson MT, Morgan GJ; UK Myeloma Forum. Ross FM, et al. Haematologica. 2010 Jul;95(7):1221-5. doi: 10.3324/haematol.2009.016329. Epub 2010 Apr 21. Haematologica. 2010. PMID: 20410185 Free PMC article. - Deregulation and Targeting of TP53 Pathway in Multiple Myeloma.
Jovanović KK, Escure G, Demonchy J, Willaume A, Van de Wyngaert Z, Farhat M, Chauvet P, Facon T, Quesnel B, Manier S. Jovanović KK, et al. Front Oncol. 2019 Jan 9;8:665. doi: 10.3389/fonc.2018.00665. eCollection 2018. Front Oncol. 2019. PMID: 30687640 Free PMC article. Review.
References
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. - PubMed
- Kyle RA, Therneau TM, Rajkumar SV, et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med. 2006;354:1362–1369. - PubMed
- Lynch HT, Watson P, Tarantolo S, et al. Phenotypic heterogeneity in multiple myeloma families. J Clin Oncol. 2005;23:685–693. - PubMed
- Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical