Detection of chromosomal structural alterations in single cells by SNP arrays: a systematic survey of amplification bias and optimized workflow - PubMed (original) (raw)
Detection of chromosomal structural alterations in single cells by SNP arrays: a systematic survey of amplification bias and optimized workflow
Kazuya Iwamoto et al. PLoS One. 2007.
Abstract
Background: In single-cell human genome analysis using whole-genome amplified product, a strong amplification bias involving allele dropout and preferential amplification hampers the quality of results. Using an oligonucleotide single nucleotide polymorphism (SNP) array, we systematically examined the nature of this amplification bias, including frequency, degree, and preference for genomic location, and we assessed the effects of this amplification bias on subsequent genotype and chromosomal copy number analyses.
Methodology/principal findings: We found a large variability in amplification bias among the amplified products obtained by multiple displacement amplification (MDA), and this bias had a severe effect on the genotype and chromosomal copy number analyses. We established optimal experimental conditions for pre-screening for high-quality amplified products, processing array data, and analyzing chromosomal structural alterations. Using this optimized protocol, we successfully detected previously unidentified chromosomal structural alterations in single cells from a lymphoblastoid cell line. These alterations were subsequently confirmed by karyotype analysis. In addition, we successfully obtained reproducible chromosomal copy number profiles of single cells from the cell line with a complex karyotype, indicating the applicability and potential of our optimized workflow.
Conclusions/significance: Our results suggest that the quality of amplification products should be critically assessed before using them for genomic analyses. The method of MDA-based whole-genome amplification followed by SNP array analysis described here will be useful for exploring chromosomal alterations in single cells.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
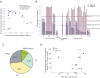
Figure 1. Visualization of the S-WGA products from a LCL by gel electrophoresis.
We loaded 1 µl of each S-WGA reaction mixture obtained by protocol 2 (N = 24). A sample indicated by an asterisk was not selected for further analysis based on the results of the Taqman genotyping assay.
Figure 2. Amplification bias and manual genotyping of the heterozygous SNP.
(A) Amplification bias revealed by the Taqman genotyping assay. The relative fluorescence intensities of alleles A and B in each sample with regard to rs1895694 are shown. Fluorescence intensities of the S-WGA products obtained by protocol 2 (N = 24) are shown. The results from another 5 heterozygous SNPs can be found in Figure S1. (B) Manual genotyping of rs1895694 in the S-WGA products. AB, heterozygous (allele AB); PA, preferential amplification; AD, allele dropout; Failed, failure in WGA. (C) Summary of the manual genotyping of the heterozygous SNPs. The percentage was calculated from a total of 240 data points obtained from genotyping of 6 heterozygous SNPs in 40 S-WGA products. (D) Concordance rate of the heterozygous SNPs by Taqman genotyping assay correlated with call rate on the SNP array. The data from 12 S-WGA products derived from a LCL and 3 S-WGA products derived from the CMK11-5 are shown. Genotyping results of 6 heterozygous SNPs (rs1895694, rs4706387, rs2074711, rs1007971, rs4140571, and rs2280964 for a LCL; rs1895694, rs7110302, rs11657541, rs1217617, rs9991, and rs2268248 for the CMK11-5) by Taqman assay were used for calculation of concordance rate. In genotyping S-WGA products, the heterozygous SNP as well as PA-classified SNPs were considered to be concordant with non-amplified gDNA. The blue squares and diamonds indicate the S-WGA products obtained by protocol 1 and 2, respectively.
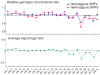
Figure 3. Concordance rate of genotypes correlated with call rate on the SNP array.
Concordance rate of genotypes was determined by the global genotype data between non-amplified gDNA and S-WGA products. The results of all genotypes (left) and homozygous/heterozygous genotypes (right) are shown. Two data sets of S-WGA products from a LCL, with call rate<50%, were not used for further analysis.
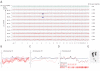
Figure 4. Genotype and CCN analyses in each chromosome.
Y axes of the top and bottom panels indicate averaged relative genotype concordance (N = 10 array) and averaged signal log2 ratio (N = 10 array), respectively. Signal log2 ratio above and below 0 indicate gain and loss of CCN, respectively. Two apparently failed array data sets (call rate<50%) were excluded from the analysis.
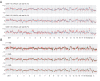
Figure 5. CCN analysis of the S-WGA products from the LCL samples.
(A) CCN profiles of S-WGA products. Standard deviations of the signal log2 ratio are shown at the right. A genomic smoothing size of 2 Mb was used for analysis. Blue bars indicate the deletion in chromosome 6q. (B) CCN profiles of chromosome 19. Blue, pink and red lines indicate CCN data of non-amplified gDNA, WGA and S-WGA products, respectively. Profiles of 10 S-WGA products are shown. (C) Example of normalization of the CCN data of chromosome 19. The CCN data of chromosome 19 in an S-WGA product was normalized using the CCN data of the other nine S-WGA products as reference. Blue, red, and black lines indicate the CCN data of non-amplified gDNA, S-WGA product, and S-WGA product (normalized), respectively. (D) Example of the deletion of chromosome 6q in the S-WGA product. Weak signal log2 ratio of the 6q was still detected after normalization was applied (black line). Red bars indicate the location of the heterozygous SNPs in the non-amplified gDNA. Heterozygous and allele dropout indicate heterozygous and homozygous calls, respectively, on the array. (E) The result of karyotyping. Deletion of chromosome 6q was detected in 4 of 14 cells analyzed.
Figure 6. CCN analysis of the S-WGA products from the CMK11-5 cell line.
(A) Results of the CCN analysis. A 3-Mb genomic smoothing size was used for analysis. Blue and red lines indicate CCN data of the non-amplified gDNA and S-WGA product, respectively. (B) Effect of the genomic smoothing size (σ) on the CCN analysis. Green and red lines indicate CCN data of the non-amplified gDNA and S-WGA product, respectively.
Similar articles
- High-resolution array copy number analyses for detection of deletion, gain, amplification and copy-neutral LOH in primary neuroblastoma tumors: four cases of homozygous deletions of the CDKN2A gene.
Carén H, Erichsen J, Olsson L, Enerbäck C, Sjöberg RM, Abrahamsson J, Kogner P, Martinsson T. Carén H, et al. BMC Genomics. 2008 Jul 29;9:353. doi: 10.1186/1471-2164-9-353. BMC Genomics. 2008. PMID: 18664255 Free PMC article. - Evaluation of genome coverage and fidelity of multiple displacement amplification from single cells by SNP array.
Ling J, Zhuang G, Tazon-Vega B, Zhang C, Cao B, Rosenwaks Z, Xu K. Ling J, et al. Mol Hum Reprod. 2009 Nov;15(11):739-47. doi: 10.1093/molehr/gap066. Epub 2009 Aug 11. Mol Hum Reprod. 2009. PMID: 19671595 Free PMC article. - Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer.
Kloth JN, Oosting J, van Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES. Kloth JN, et al. BMC Genomics. 2007 Feb 20;8:53. doi: 10.1186/1471-2164-8-53. BMC Genomics. 2007. PMID: 17311676 Free PMC article. - Multiple displacement amplification to create a long-lasting source of DNA for genetic studies.
Lovmar L, Syvänen AC. Lovmar L, et al. Hum Mutat. 2006 Jul;27(7):603-14. doi: 10.1002/humu.20341. Hum Mutat. 2006. PMID: 16786504 Review. - SNP array analysis in constitutional and cancer genome diagnostics--copy number variants, genotyping and quality control.
de Leeuw N, Hehir-Kwa JY, Simons A, Geurts van Kessel A, Smeets DF, Faas BH, Pfundt R. de Leeuw N, et al. Cytogenet Genome Res. 2011;135(3-4):212-21. doi: 10.1159/000331273. Epub 2011 Sep 16. Cytogenet Genome Res. 2011. PMID: 21934286 Review.
Cited by
- A Method for Detection of Somatic LINE-1 Insertions at the Single-Cell Level from Postmortem Human Brain.
Bundo M, Iwamoto K. Bundo M, et al. Methods Mol Biol. 2023;2577:147-159. doi: 10.1007/978-1-0716-2724-2_10. Methods Mol Biol. 2023. PMID: 36173571 - Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells.
Babayan A, Alawi M, Gormley M, Müller V, Wikman H, McMullin RP, Smirnov DA, Li W, Geffken M, Pantel K, Joosse SA. Babayan A, et al. Oncotarget. 2016 Jul 19;8(34):56066-56080. doi: 10.18632/oncotarget.10701. eCollection 2017 Aug 22. Oncotarget. 2016. PMID: 28915574 Free PMC article. - Number of blastocysts biopsied as a predictive indicator to obtain at least one normal/balanced embryo following preimplantation genetic diagnosis with single nucleotide polymorphism microarray in translocation cases.
Wang YZ, Ding CH, Wang J, Zeng YH, Zhou W, Li R, Zhou CQ, Deng MF, Xu YW. Wang YZ, et al. J Assist Reprod Genet. 2017 Jan;34(1):51-59. doi: 10.1007/s10815-016-0831-0. Epub 2016 Nov 7. J Assist Reprod Genet. 2017. PMID: 27822654 Free PMC article. - Direct squencing from the minimal number of DNA molecules needed to fill a 454 picotiterplate.
Džunková M, Garcia-Garcerà M, Martínez-Priego L, D'Auria G, Calafell F, Moya A. Džunková M, et al. PLoS One. 2014 Jun 2;9(6):e97379. doi: 10.1371/journal.pone.0097379. eCollection 2014. PLoS One. 2014. PMID: 24887077 Free PMC article. - Reliable single cell array CGH for clinical samples.
Czyż ZT, Hoffmann M, Schlimok G, Polzer B, Klein CA. Czyż ZT, et al. PLoS One. 2014 Jan 21;9(1):e85907. doi: 10.1371/journal.pone.0085907. eCollection 2014. PLoS One. 2014. PMID: 24465780 Free PMC article.
References
- Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. - PubMed
- Iourov IY, Vorsanova SG, Yurov YB. Chromosomal variation in mammalian neuronal cells: known facts and attractive hypotheses. Int Rev Cytol. 2006;249:143–191. - PubMed
- Lasken RS, Egholm M. Whole genome amplification: abundant supplies of DNA from precious samples or clinical specimens. Trends Biotechnol. 2003;21:531–535. - PubMed
- Lovmar L, Syvanen AC. Multiple displacement amplification to create a long-lasting source of DNA for genetic studies. Hum Mutat. 2006;27:603–614. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources