The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae - PubMed (original) (raw)
The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae
Heesun Cheong et al. Mol Biol Cell. 2008 Feb.
Abstract
Autophagy is the major degradative process for recycling cytoplasmic constituents and eliminating unnecessary organelles in eukaryotic cells. Most autophagy-related (Atg) proteins are recruited to the phagophore assembly site (PAS), a proposed site for vesicle formation during either nonspecific or specific types of autophagy. Therefore, appropriate recruitment of Atg proteins to this site is critical for their function in autophagy. Atg11 facilitates PAS recruitment for the cytoplasm-to-vacuole targeting pathway, which is a specific, autophagy-like process that occurs under vegetative conditions. In contrast, it is not known how Atg proteins are recruited to the PAS, nor which components are involved in PAS formation under nonspecific autophagy-inducing, starvation conditions. Here, we studied PAS assembly during nonspecific autophagy, using an atg11Delta mutant background to eliminate the PAS formation that occurs during vegetative growth. We found that protein complexes containing the Atg1 kinase have two roles for PAS formation during nonspecific autophagy. The Atg1 C terminus mediates an interaction with Atg13 and Atg17, facilitating a structural role of Atg1 that is needed to efficiently organize an initial step of PAS assembly, whereas Atg1 kinase activity affects the dynamics of protein movement at the PAS involved in Atg protein cycling.
Figures
Figure 1.
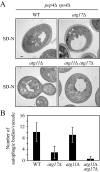
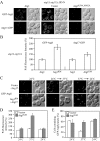
Atg17 is important for PAS organization under starvation conditions. Wild-type (SEY6210), _atg11_Δ (AHY001), _atg17_Δ (CWY239), and _atg11_Δ _atg17_Δ (HCY66) strains expressing GFP-Atg8 or GFP-Atg1 from centromeric plasmids were grown in SMD lacking auxotrophic amino acids and shifted to SD-N for 2 h before microscopy. The cells were examined by fluorescence microscopy as described in Materials and Methods.
Figure 2.
The _atg11_Δ _atg17_Δ double mutant is defective for autophagosome formation. (A) The wild-type (FRY143; pep4Δ _vps4_Δ), _atg17_Δ (HCY31; pep4Δ _vps4_Δ), _atg11_Δ (HCY37; pep4Δ _vps4_Δ), and _atg11_Δ _atg17_Δ (HCY38; pep4Δ _vps4_Δ) strains were grown to mid-log stage in YPD and transferred to SD-N medium for 4 h. Cells were fixed with permanganate and examined by electron microscopy as described in Materials and Methods. (B) Quantification of autophagic body accumulation. 50 sections for each strain were scored for autophagic body accumulation.
Figure 3.
Atg13 and Atg1 are required for PAS recruitment of Atg proteins under starvation conditions. Localization of GFP-Atg8 (A) or GFP-Atg1 (B) in _atg11_Δ (AHY001), _atg13_Δ (CWY233), _atg1_Δ _atg11_Δ (TYY158), and _atg11_Δ _atg13_Δ (CWY235) strains, and GFP-Atg17 (C) in the _atg1_Δ _atg11_Δ (TYY158) strain was examined by fluorescence microscopy as described in Figure 1.
Figure 4.
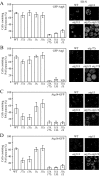
Atg1 and its modulators Atg13 and Atg17 are required for the PAS recruitment of various Atg proteins under starvation conditions. The localization of Atg proteins was examined by fluorescence microscopy as described in Figure 1. The number of cells that contained PAS puncta was quantified for GFP-Atg8 (A), GFP-Atg1 (B), Atg14-GFP (C), and Atg16-GFP (D), after shifting to SD-N for 2 h. Approximately 150–250 cells for each strain were analyzed for scoring the percentage of cells with fluorescent PAS puncta. ND, not determined.
Figure 5.
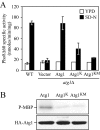
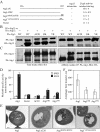
Atg1 kinase activity is required for autophagy. (A) Atg1 mutants are defective for autophagy. The alkaline phosphatase activity from Pho8Δ60, a marker for nonspecific autophagy, was monitored to determine the level of autophagy activity. The wild-type (YTS159) and _atg1_Δ (UNY1) strains harboring plasmids expressing Atg1, Atg1K54A (Atg1K), Atg1K54A,M102A (Atg1KM), or empty vector (pRS415) were grown in SMD lacking auxotrophic amino acids and shifted to SD-N for 4 h. Samples were collected and protein extracts assayed for alkaline phosphatase activity as described in Materials and Methods. The results represent the mean and SD of three experiments. (B) Atg1 mutants are defective for kinase activity. The _atg1_Δ mutant (WHY001) cells expressing HA-Atg1, HA-Atg1K54A, or HA-Atg1K54A,M102A were treated with 0.2 μg/ml rapamycin for 2 h. The extract of each mutant was immunoprecipitated with anti-HA antibody, and then immunocomplexes were assayed for Atg1 kinase activity as described in Materials and Methods.
Figure 6.
Atg1 kinase activity is required for autophagosome formation. (A) Atg1 mutants are defective for autophagic body accumulation. The _atg1_Δ (HCY76; _pep4_Δ _vps4_Δ) strains harboring plasmids expressing Atg1, Atg1K54A (Atg1K), Atg1K54A,M102A (Atg1KM), or the empty vector (pRS415) were grown in selective medium and shifted to SD-N for 4 h. Cells were fixed with permanganate and examined by electron microscopy as described in Materials and Methods. (B) Quantification of autophagic body accumulation. Fifty sections for each strain were scored for autophagic body accumulation in the vacuole.
Figure 7.
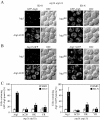
Atg1 kinase activity is not essential for PAS localization of Atg8 and Atg17, but it is required for normal localization of these proteins under starvation conditions. (A) Cells from the _atg1_Δ _atg11_Δ strain expressing GFP-Atg8 (HCY116) and Atg17-3xGFP (HCY119) from the respective chromosomal loci were transformed with plasmids encoding Atg1, empty vector, or Atg1K54A,M102A. The cells were grown in SMD and shifted to SD-N for 2 h before fluorescence microscopy, which was performed as described in Materials and Methods. (B) For quantification of fluorescence intensity at the PAS, 30 individual cells for each strain in A were observed by fluorescence microscopy. The relative percentage of fluorescence intensity was calculated as described in Materials and Methods and normalized to the value for wild-type Atg1 cells, which were set at 100%. Error bars indicate the SD of three independent experiments. (C) Redistribution of Atg8 from the PAS is affected by Atg1 kinase activity under starvation conditions. The _atg1_Δ _atg11_Δ strain expressing GFP-Atg8 from the chromosomal locus (HCY116) was cotransformed with plasmids encoding ATG1 ts and Atg1K54A,M102A or ATG1 ts and empty vector. The cells were grown in SMD medium at 24°C and transferred to SD-N for 1 h before the temperature shift. For inactivation of Atg1ts, cells were shifted to 37°C for 2 h, and then shifted back to 24°C for 2 h. The cells were examined by fluorescence microscopy as described in Materials and Methods. (D) For quantification of fluorescence intensity at the PAS, 30 individual cells from each strain in C were observed by fluorescence microscopy. The relative percentage of fluorescence intensity was calculated and normalized based on the value for GFP-Atg8 in cells expressing Atg1ts and the empty vector, which was set at 100% at the permissive temperature. Error bars indicate the SD of three independent experiments. (E) Quantification of the number of cells that contained GFP-Atg8 puncta as a PAS marker from C. Approximately 150–200 cells for each strain were analyzed for scoring the percentage of cells containing fluorescent dots.
Figure 8.
Atg1 kinase activity is important for Atg8 dynamics during starvation. Cells from the _atg1_Δ _atg11_Δ strain expressing GFP-Atg8 (HCY116) were transformed with plasmids encoding Atg1 and Atg1K54A,M102A (Atg1KM). The cells were grown in SMD medium and shifted to SD-N for 2 h before fluorescence microscopy. For time-lapse experiments, the cells were immobilized on SD-N medium containing 2% agar on concavity slides as described in Materials and Methods, and pictures were taken every minute. Images are shown over the 7-min time course.
Figure 9.
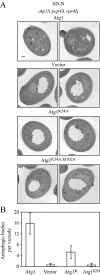
The interaction between Atg1 and Atg13 is important for autophagy. (A) Mutations in the C terminus of Atg1 disrupt its interaction with Atg13 based on a yeast two-hybrid assay. PJ69-4A cells expressing Atg13 (AD-Atg13) and either Atg1 lacking the N-terminal kinase domain (BD-Atg1ΔN), Atg1ΔN deleted for the C-terminal 20 amino acids (BD-Atg1ΔC20), Atg1ΔN with substitutions of R885E and K892E (BD-Atg1R885E,K892E), Atg1ΔN with substitutions of Y878A and R885A (BD-Atg1Y878A,R885A), or the BD-empty vector were grown for 5 d on plates lacking adenine. Growth or lack of growth on these plates was scored as + or −, respectively. The strength of the interaction for each set of proteins was quantified by measuring β-galactosidase activity from three independent experiments. (B and C) Atg1 C-terminal mutants are defective for interacting with Atg13 but not Atg11. The B, _atg1_Δ _atg13_Δ (UNY27) or C, _atg1_Δ _atg11_Δ (YTS157) cells were transformed with a plasmid expressing 3xHA-Atg13 under the control of its endogenous promoter or 3xHA-Atg11 under the control of the CUP1 promoter as indicated, and CUP1 promoter-driven protein A (PA)-tagged fusions of either wild-type Atg1 (WT), Atg1ΔC20 (ΔC20), Atg1R885E,K892E (RK), or Atg1Y878A,R885A (YR). As negative controls UNY27 and YTS157 cells were transformed with a plasmid expressing CUP1 promoter-driven wild-type PA-Atg1 and an empty pRS315 or pRS414 plasmid, respectively. Protein extracts were subjected to affinity isolation, eluted proteins were separated on a 6% SDS-PAGE gel and detected with anti-HA antibody as described in Materials and Methods. IP, immunoaffinity-purified isolate. (D) The Atg1 C-terminal mutants are defective in autophagy. The Pho8Δ60 assay was used to monitor autophagy activity for the _atg1_Δ (UNY3) strain expressing the indicated wild-type Atg1, empty vector (pRS315), or Atg1 mutant plasmids expressing Atg1 under the control of the endogenous promoter, as described in Materials and Methods. Alkaline phosphatase activity was monitored from protein extracts prepared from cells grown in SMD or after a 4 h shift to SD-N. The results represent the mean and SD of three independent experiments. (E) The Atg1ΔC20 mutant is defective in forming autophagosomes (detected as autophagic bodies), whereas small autophagosomes are formed in cells expressing C-terminal point mutants. An _atg1_Δ _pep4_Δ _vps4_Δ strain (HCY76) was transformed individually with empty vector, or plasmids expressing wild-type Atg1 or the indicated Atg1 mutant. Cells were grown to mid-log phase in SMD medium, shifted to SD-N medium to induce starvation, fixed with potassium permanganate, and examined by electron microscopy as described in Materials and Methods. No autophagic bodies were detected in cells transformed with the empty vector. (F) Quantification of the diameters of the autophagic bodies was carried out from 50 sections for each strain. Essentially no autophagic bodies were detected in the strain expressing Atg1ΔC20.
Figure 10.
The interaction of Atg1 with Atg13 is required for PAS formation under starvation conditions. Cells from the _atg1_Δ _atg11_Δ strain expressing GFP-Atg8 (HCY116) (A) or Atg17-GFP (HCY119) (B) from the chromosomal locus were transformed with plasmids encoding wild-type Atg1, or the Atg1ΔC20 (ΔC20), Atg1R885E,K892E (RK), or Atg1 Y878A,R885A (YR) mutants. The cells were grown in SMD and shifted to SD-N for 2 h before fluorescence microscopy, which was carried out as described in Materials and Methods. (C) Quantification of the number of cells that contained GFP-Atg8 or Atg17-GFP puncta as a PAS marker from A and B. Approximately 150–200 cells for each strain were analyzed for scoring the percentage of cells with fluorescent dots.
Similar articles
- Dual role of Atg1 in regulation of autophagy-specific PAS assembly in Saccharomyces cerevisiae.
Cheong H, Klionsky DJ. Cheong H, et al. Autophagy. 2008 Jul;4(5):724-6. doi: 10.4161/auto.6375. Epub 2008 Jun 2. Autophagy. 2008. PMID: 18552550 - Atg17 recruits Atg9 to organize the pre-autophagosomal structure.
Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Sekito T, et al. Genes Cells. 2009 May;14(5):525-38. doi: 10.1111/j.1365-2443.2009.01299.x. Epub 2009 Apr 13. Genes Cells. 2009. PMID: 19371383 - Organization of the pre-autophagosomal structure responsible for autophagosome formation.
Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Kawamata T, et al. Mol Biol Cell. 2008 May;19(5):2039-50. doi: 10.1091/mbc.e07-10-1048. Epub 2008 Feb 20. Mol Biol Cell. 2008. PMID: 18287526 Free PMC article. - Mechanistic Insights into the Role of Atg11 in Selective Autophagy.
Zientara-Rytter K, Subramani S. Zientara-Rytter K, et al. J Mol Biol. 2020 Jan 3;432(1):104-122. doi: 10.1016/j.jmb.2019.06.017. Epub 2019 Jun 22. J Mol Biol. 2020. PMID: 31238043 Free PMC article. Review. - The Atg17-Atg31-Atg29 Complex Coordinates with Atg11 to Recruit the Vam7 SNARE and Mediate Autophagosome-Vacuole Fusion.
Liu X, Mao K, Yu AYH, Omairi-Nasser A, Austin J 2nd, Glick BS, Yip CK, Klionsky DJ. Liu X, et al. Curr Biol. 2016 Jan 25;26(2):150-160. doi: 10.1016/j.cub.2015.11.054. Epub 2016 Jan 7. Curr Biol. 2016. PMID: 26774783 Free PMC article. Review.
Cited by
- AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis.
Li F, Chung T, Vierstra RD. Li F, et al. Plant Cell. 2014 Feb;26(2):788-807. doi: 10.1105/tpc.113.120014. Epub 2014 Feb 21. Plant Cell. 2014. PMID: 24563201 Free PMC article. - DeepPhagy: a deep learning framework for quantitatively measuring autophagy activity in Saccharomyces cerevisiae.
Zhang Y, Xie Y, Liu W, Deng W, Peng D, Wang C, Xu H, Ruan C, Deng Y, Guo Y, Lu C, Yi C, Ren J, Xue Y. Zhang Y, et al. Autophagy. 2020 Apr;16(4):626-640. doi: 10.1080/15548627.2019.1632622. Epub 2019 Jun 20. Autophagy. 2020. PMID: 31204567 Free PMC article. - The independence of and associations among apoptosis, autophagy, and necrosis.
Chen Q, Kang J, Fu C. Chen Q, et al. Signal Transduct Target Ther. 2018 Jul 1;3:18. doi: 10.1038/s41392-018-0018-5. eCollection 2018. Signal Transduct Target Ther. 2018. PMID: 29967689 Free PMC article. - Anatomy of autophagy: from the beginning to the end.
Zhi X, Feng W, Rong Y, Liu R. Zhi X, et al. Cell Mol Life Sci. 2018 Mar;75(5):815-831. doi: 10.1007/s00018-017-2657-z. Epub 2017 Sep 22. Cell Mol Life Sci. 2018. PMID: 28939950 Free PMC article. Review. - The Atg1 complex, Atg9, and Vac8 recruit PI3K complex I to the pre-autophagosomal structure.
Hitomi K, Kotani T, Noda NN, Kimura Y, Nakatogawa H. Hitomi K, et al. J Cell Biol. 2023 Aug 7;222(8):e202210017. doi: 10.1083/jcb.202210017. Epub 2023 Jul 12. J Cell Biol. 2023. PMID: 37436710 Free PMC article.
References
- Birmingham C. L., Brumell J. H. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006;2:156–158. - PubMed
- Colombo M. I., Gutierrez M. G., Romano P. S. The two faces of autophagy: Coxiella and Mycobacterium. Autophagy. 2006;2:162–164. - PubMed
- Dunn W. A., Jr, Cregg J. M., Kiel J.A.K.W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials