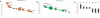A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells - PubMed (original) (raw)
A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells
Ingrid Ibarra et al. Genes Dev. 2007.
Abstract
microRNA (miRNA) expression profiles are often characteristic of specific cell types. The mouse mammary epithelial cell line, Comma-Dbeta, contains a population of self-renewing progenitor cells that can reconstitute the mammary gland. We purified this population and determined its miRNA signature. Several microRNAs, including miR-205 and miR-22, are highly expressed in mammary progenitor cells, while others, including let-7 and miR-93, are depleted. Let-7 sensors can be used to prospectively enrich self-renewing populations, and enforced let-7 expression induces loss of self-renewing cells from mixed cultures.
Figures
Figure 1.
Characterization of ALDH as a marker for progenitor cells in Comma-Dβ. (A) A FACS pseudo-color dot plot showing ALDH activity and Sca-1 expression in Comma-Dβ cells. (Left) Cells incubated with ALDEFLOUR substrate and stained with Sca-1. (Right) Cells stained with ALDEFLOUR and costained with Sca-1 and incubated with DEAB to establish background fluorescence. Shown are 100,000 events. (B) Histogram showing the colony-forming capacity of four sorted populations based on ALDH activity and Sca-1 expression seeded at clonal density on irradiated NIH3T3 feeders. Data represent the mean of four independent experiments. (C) Giemsa staining of ALDHbrScahi colonies grown on irradiated feeders for 6 d. Based on morphology, myoepithelial (top), luminal (middle), and mixture (bottom) colonies were observed. (D) Histogram showing the colony-forming capacity of four sorted populations embedded at clonal density in Matrigel (n = 4). (E) FACS profile of Comma-Dβ cells treated with a 6 μM dose of MAF for 4 d. (Left) Cells incubated with ALDEFLOUR and stained with Sca-1. (Right) DEAB control. (F) Cell viability assay after a 24-h treatment with various doses of MAF of Comma-Dβ cells (black) and MAF-resistant cells (blue). Data represent the mean ± SD (error bar) of two independent experiments done in triplicate.
Figure 2.
MiRNAs are differentially expressed in self-renewing compartments. (A,B) Bubble plots depicting the relative abundance and log2 ratio of miRNAs in ALDHbrScahi (A) and MAF-resistant (B) cells relative to Sca-1negative cells. (C) qRT–PCR for the mature forms of selected differentially expressed miRNAs. Shown are relative expression levels ΔΔCT of each miRNA from sorted Sca-1hi and Sca-1neg Comma-Dβ cells.
Figure 3.
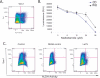
let-7c depletes the self-renewing compartment in Comma-Dβ. (A) Ectopic expression of Wnt expands the ALDHbr Sca-1hi compartment. FACS plot of Comma-Dβ overexpressing Wnt-1 costained with ALDEFLOUR and Sca-1. (B) Cell viability assay after a 48-h treatment with various doses of MAF of Comma-Dβ cells (blue) and Wnt-1-expressing cells (black). Data represent the mean ± SD (error bar) of two independent experiments done in triplicate. (C) FACS profile of empty vector control Comma-Dβ cells costained with ALDEFLOUR and Sca-1 (left), DEAB control for empty vector cells (middle), and Comma-Dβ cells ectopically expressing Let7c (right) and also stained with ALDEFLOUR and Sca-1. A depletion of the ALDH compartment is observed upon introduction of Let7c.
Figure 4.
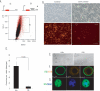
Self-renewal and differentiation of let-7c-negative cells in vitro. (A) Cartoon depicting the let-7c sensor construct. (B) Phase-contrast images of Comma-Dβ cells expressing a construct with no let-7c-binding sites (control) and Comma-Dβ cells expressing a sensor construct containing let-7c complementary sites. (C) Overlay FACS dot plot of let-7c sensor cells (red) and uninfected Comma-Dβ cells (black) as an unstained control. DsR+ cells constitute 0.8% of the total population. (D) Histogram showing the colony-forming ability of DsR+ and DsR− cells embedded at clonal density in Matrigel (n = 4). (E) Phase-contrast images of resultant DsR+ and DsR− spheroids grown on Matrigel. DsR+ cells gave rise to substantially larger colonies (>50 μm), whereas DsR− cells never exceeded this size. (F,G) Confocal images of spheroids derived from DsR+ cells. (F) Representative cross-section through the middle of a sphere costained with basal K5 and luminal K18 antibodies. (G) Representative image through the top of a spheroid costained with basal K5 and α-Sma antibodies.
Similar articles
- A putative role for microRNA-205 in mammary epithelial cell progenitors.
Greene SB, Gunaratne PH, Hammond SM, Rosen JM. Greene SB, et al. J Cell Sci. 2010 Feb 15;123(Pt 4):606-18. doi: 10.1242/jcs.056812. Epub 2010 Jan 26. J Cell Sci. 2010. PMID: 20103531 Free PMC article. - Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dbeta cell line.
Deugnier MA, Faraldo MM, Teulière J, Thiery JP, Medina D, Glukhova MA. Deugnier MA, et al. Dev Biol. 2006 May 15;293(2):414-25. doi: 10.1016/j.ydbio.2006.02.007. Epub 2006 Mar 20. Dev Biol. 2006. PMID: 16545360 - The proto-oncogene Myc is essential for mammary stem cell function.
Moumen M, Chiche A, Deugnier MA, Petit V, Gandarillas A, Glukhova MA, Faraldo MM. Moumen M, et al. Stem Cells. 2012 Jun;30(6):1246-54. doi: 10.1002/stem.1090. Stem Cells. 2012. PMID: 22438054 - Delineating the epithelial hierarchy in the mouse mammary gland.
Asselin-Labat ML, Vaillant F, Shackleton M, Bouras T, Lindeman GJ, Visvader JE. Asselin-Labat ML, et al. Cold Spring Harb Symp Quant Biol. 2008;73:469-78. doi: 10.1101/sqb.2008.73.020. Epub 2008 Nov 6. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19022771 Review. - Does the Mouse Mammary Gland Arise from Unipotent or Multipotent Mammary Stem/Progenitor Cells?
Smith GH, Medina D. Smith GH, et al. J Mammary Gland Biol Neoplasia. 2018 Jun;23(1-2):1-3. doi: 10.1007/s10911-018-9394-2. Epub 2018 Apr 11. J Mammary Gland Biol Neoplasia. 2018. PMID: 29644495 Review.
Cited by
- Mechanistic insights into the role of microRNAs in cancer: influence of nutrient crosstalk.
Shah MS, Davidson LA, Chapkin RS. Shah MS, et al. Front Genet. 2012 Dec 28;3:305. doi: 10.3389/fgene.2012.00305. eCollection 2012. Front Genet. 2012. PMID: 23293655 Free PMC article. - MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling.
Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, Richardson AL, Pandolfi PP. Song SJ, et al. Cell. 2013 Jul 18;154(2):311-324. doi: 10.1016/j.cell.2013.06.026. Epub 2013 Jul 3. Cell. 2013. PMID: 23830207 Free PMC article. - MiR-15a decreases bovine mammary epithelial cell viability and lactation and regulates growth hormone receptor expression.
Li HM, Wang CM, Li QZ, Gao XJ. Li HM, et al. Molecules. 2012 Oct 12;17(10):12037-48. doi: 10.3390/molecules171012037. Molecules. 2012. PMID: 23085654 Free PMC article. - The role of non-coding RNAs in the regulation of stem cells and progenitors in the normal mammary gland and in breast tumors.
Tordonato C, Di Fiore PP, Nicassio F. Tordonato C, et al. Front Genet. 2015 Feb 27;6:72. doi: 10.3389/fgene.2015.00072. eCollection 2015. Front Genet. 2015. PMID: 25774169 Free PMC article. Review. - Prostate cancer stem cell biology.
Yu C, Yao Z, Jiang Y, Keller ET. Yu C, et al. Minerva Urol Nefrol. 2012 Mar;64(1):19-33. Minerva Urol Nefrol. 2012. PMID: 22402315 Free PMC article. Review.
References
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Elledge S.J., Anderson K.V., Hannon G.J., Anderson K.V., Hannon G.J., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. - PubMed
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. - PMC - PubMed
- Bunting K.D., Townsend A.J., Townsend A.J. De novo expression of transfected human class 1 aldehyde dehydrogenase (ALDH) causes resistance to oxazaphosphorine anti-cancer alkylating agents in hamster V79 cell lines. Elevated class 1 ALDH activity is closely correlated with reduction in DNA interstrand cross-linking and lethality. J. Biol. Chem. 1996;27:11884–11890. - PubMed
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Xu N.L., Mahuvakar V.R., Andersen M.R., Mahuvakar V.R., Andersen M.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical