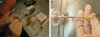Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection - PubMed (original) (raw)
Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection
Jason B Bell et al. Nat Protoc. 2007.
Abstract
Nonviral, DNA-mediated gene transfer is an alternative to viral delivery systems for expressing new genes in cells and tissues. The Sleeping Beauty (SB) transposon system combines the advantages of viruses and naked DNA molecules for gene therapy purposes; however, efficacious delivery of DNA molecules to animal tissues can still be problematic. Here we describe the hydrodynamic delivery procedure for the SB transposon system that allows efficient delivery to the liver in the mouse. The procedure involves rapid, high-pressure injection of a DNA solution into the tail vein. The overall procedure takes <1 h although the delivery into one mouse requires only a few seconds. Successful injections result in expression of the transgene in 5-40% of hepatocytes 1 d after injection. Several weeks after injection, transgene expression stabilizes at approximately 1% of the level at 24 h, presumably owing to integration of the transposons into chromosomes.
Figures
Figure 1
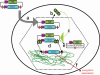
Schematic of Sleeping Beauty (SB) transposition. DNA transposition consists of a cut-and-paste reaction in which a transposon containing a gene of interest (GOI, shown in blue, with its promoter shown in magenta) is cut out of a plasmid and inserted into another DNA molecule, in this case a mouse chromosome. The cleavage reaction occurs at the ends of the inverted terminal repeats (inverted set of double arrowheads in the diagram) of the SB transposon. SB transposons integrate only into TA-dinucleotide basepairs (∼200 million in mammalian genomes). The inverted terminal repeats are the only DNA sequences required by the transposase enzyme for transposition. The transposase gene can be on the same plasmid (cis configuration as shown) or delivered on a separate plasmid (trans configuration, see Fig. 3) to permit greater flexibility in experimental design. (a) The plasmid carrying the SB transposase gene and transposon enters a cell (large back oval) as a result of hydrodynamic delivery and proceeds through the nuclear membrane (dotted hexagon) by a poorly understood process. (b) The SB transposase gene (SB, green) is transcribed from a promoter (magenta arrowhead) and the mRNA translated in the cytoplasm to give an appropriate level of enzyme (green circles). (c) The SB transposase molecules enter the nucleus and bind to the transposon, two at each end, to bring the transposon ends together (shown in detail in Fig. 3b). (d) Four transposase enzymes work in concert to cleave the plasmid at the termini of the transposon and paste it (dotted lines) into chromosomal DNA (green tangled lines). (e) A plasmid excision product is left behind in this reaction (the site whence the transposon left is marked by an X). (f) Integration into a chromosome can confer long-term expression of the GOI that is contained within the transposon.
Figure 2
Excision assay. (a) Theory of the excision assay. The first step of transposition is precise cleavage on both sides of the transposon (excision step, step d in Fig. 1). The gap in the donor-plasmid left by the transposon is repaired (step e in Fig. 1) to reform a circular plasmid lacking the transposon. The re-ligation step often leaves a footprint of an extra 5 bp (either TCTGA or TCAGA) at the site of the original transposon. As a result of ligation of the plasmid, primers (red arrows in the bottom right corner of panel a) complementary to plasmid sequences located on either side of the site whence the transposon originated can bind to the repaired plasmid. PCR-amplification from the primers leads to a DNA sequence of predictable length and sequence (except for the footprint), which we refer to as the excision product (EP) (red line at the bottom right of panel a). The power of the assay is that regardless of where the transposon integrates, the EP from each event is the same thereby allowing quantification of the totality of transposition events. Thus, whereas identification of integration at any of 2 × 108 TA sites can only be determined through analysis of genomic DNA, the intensity of the PCR product from the repaired plasmid can be used to access the overall extent of transposition. The red line represents the EP from PCR and the red arrow indicates its location on the gel in panel c. Primer sites are chosen to be 100−200 bp outside of the inverted repeats of the transposon to ensure that the PCR product is efficiently synthesized, that the EP is distinguishable from the primers (and ‘primer dimers’ sometimes seen in gels of PCRs), and that the EP is distinguishable from the PCR amplification across the transposon from unexcised plasmids. (b) The level of transposition can be estimated through conventional PCR and analysis by agarose-gel electrophoresis. The gel shows a single band amplified from a genomic sequence (e.g., β-glucuronidase) in a liver sample from a sham-treated control mouse that received an injection of lactated Ringer's solution without DNA and an additional band representing the amplified EP (red EP) from a mouse injected with a transposon-containing plasmid. (c) The amount of EP can be quantitatively determined by real-time PCR using any of several instruments (the data here were generated using an iCycler (Bio-Rad Laboratories) with their IQ SYBR green supermix according to their guidelines). The data show the copy numbers of excision plasmid product (EP) as a function of the PCR threshold cycle (_C_t) number when SYBR Green fluorescence is first detectable. The standard curve indicates that the signal is proportional to the input level of repaired plasmids after excision (open boxes). Triplicate samples were assayed and overlapped for all samples except that from the sham-treated mouse (two triangles; the bottom triangle represents two results) and the untreated mouse (black circles with two of the samples overlapping at the bottom). Greater variation between samples is sometimes seen with the least concentrated samples because the slightest contamination can lead to a measurable signal. The gel at the top shows the final products from real-time PCRs that were resolved by electrophoresis using 1% agarose gel. Panel a is adapted from Figure 2 of Liu et al. (2004) and panels b and c are from Figure 6 of Aronovich et al. (2007). GOI, gene of interest; SB, Sleeping Beauty.
Figure 3
Design of Sleeping Beauty (SB) transposons. (a) The cis and trans configurations for delivery of the SB system are shown with transcriptional regulatory sequences represented by the magenta arrowheads as described in Figure 1. In the _trans_-delivery mode, the transposase gene can be omitted as a negative control. An alternative _trans_-delivery is possible wherein the SB mRNA is delivered rather than its gene. (b) The phenomenon of overproduction inhibition is illustrated. The rate of transposition depends on the binding of four transposase enzymes to the transposon [SB]optimal. If there are fewer than four transposase molecules [SB]low, transposition will not occur and if there are more, then the additional transposase molecules will quench the reaction by competing with bound enzymes to prevent the bringing of the transposon ends together [SB]high. (c) The effects of cargo (expression cassette) size on gene delivery as determined by long-term gene expression in dividing cells. The cause for the apparent decrease in transposition as a function of size is poorly understood because the length of the plasmid carrier sets the linear distance between the termini of the transposon once the expression cassette is longer than ∼2 kb. Factors that may influence the efficiency of gene delivery include propagation through the plasma and nuclear membranes, where effects of DNA length have not been carefully evaluated. The diagrams in panels b and c are adapted, with permission, from Figure 3 of Geurts et al. (2003) and Figure 2 of Hackett (2007),respectively.
Figure 4
Hydrodynamic injection procedure of a transposon containing a gene of interest and a source of Sleeping Beauty (SB) transposase encoded by an SB gene. The components of the SB system are the same as in Figures 1 and 3. _Trans_- and _cis_-deliveries are performed in the same way. (a) The desired amount of DNA (generally between 0.1 and 50 μg, although higher and lower amounts can be injected) is diluted into a volume of lactated Ringer's solution that is equivalent to 10% the mouse weight. (b) The DNA solution is injected into the tail vein of the mouse; injections that take 4−7 s are optimal. (c) Results from hydrodynamic delivery can be obtained as soon as 30 min after injection, depending on the gene of interest (GOI) and the assay.
Figure 5
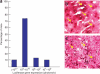
Hydrodynamic delivery to liver. (a) The graph shows the reproducibility of delivery of a luciferase expression cassette. Seventy-five percent of the C57BL/6 mice express a luciferase transgene within a tenfold range (2 × 109 to 2 × 1010 relative light units). (b) The image shows the cellular distribution of expression of human β-glucuronidase (dark purple cells) in liver sections from a C57BL/6 mouse 24 h after hydrodynamic injection of transposons carrying the gene controlled by a hybrid cytomegalovirus (CMV)/β-actin promoter (yellow arrows show some of the cells expressing β-glucuronidase). (c) The image shows the cellular distribution of expression of human β-glucuronidase (dark purple cells) in liver sections from a β-glucuronidase-deficient C57BL/6 mouse 24 h after injection following hydrodynamic injection of the same transposon vector as in panel b. All of the cells are stained because those that do not express the enzyme take up enzyme produced by those cells that are expressing the transgene, a process called ‘cross-correction’. Panel c is adapted from Figure 1 of Aronovich et al. (2007).
Figure 6
Layout of equipment for hydrodynamic injection. The items, except for a marker pen in middle are numbered for easy identification: 1) Bleach bucket, for washing items and hands that are used in the hood in accordance with specific-pathogen free (SPF) procedure. 2) Bleach Spray bottle to facilitate washing in accordance with SPF procedure. 3) Lactated Ringer's solution for dilution and injection of transgenic DNA. 4) Three milliliter syringes for hydrodynamic injections. 5) Butterfly needles for hydrodynamic injections; the needle attaches to the 3-ml syringe. 6) Rack with samples that contain the DNA solutions for injection. 7) One-milliliter syringe with anesthetic drug already drawn up; it is covered because the anesthetic is sensitive to visible light. 8) Alcohol pads to sanitize tails. 9) Timer to measure duration of each injection. 10) Scale for weighing mice. 11) Mouse restrainer to contain mouse and reduce its activity during the injection. 12) Top of mouse cage set at an angle for better viewing. 13) Bottom of mouse cage for animal housing. 14) Heat lamp and ring stand to dilate veins in mice and keep them warm. The vertical rod in the stand is ∼50 cm tall.
Figure 7
Hydrodynamic injection with mouse in restrainer. (a) The mouse is placed in a restrainer and the tail is rotated until the lateral vein faces up. In this example, the needle is in the right hand and is being placed into the tail vein for injection. (b) Detail of insertion of the needle into a tail vein. Two tail arteries run along the top and the bottom of the tail and two tail veins run on either side, midway between the arteries. The needle is inserted ∼2−3 cm from the tip of the tail (which is under the injector's thumb in this image). The needle is almost completely inserted into the vein for best results. The tip of the tail behind the injection site may be bent downward by some injectors. The tail is pulled taut, but excessive force may result in the tail coming off in young mice. If the injection fails, a new site, roughly in the middle of the tail can be tried. If this fails, one can try the other tail vein on the other side. When the needle is removed, some blood will flow from the puncture; bleeding is stopped by pressing lightly (not squeezing tightly) the wound with a finger and thumb.
Figure 8
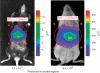
Typical results from in vivo bioluminescence imaging of mice 24 h after hydrodynamic injection. Both mice are on black construction paper, which may appear differently according to background illumination. Color-coded luminescence scales for the circled regions are shown on the right. Exposures are 1 s. The imaging identifies the liver as the primary site of expression of a luciferase expression cassette. The mouse on the left is a C57BL/6 mouse (Imaged for 0.5 s) and the mouse on the right is a NOD.129(B6)-PrkdcscidIduatm1Clk (imaged for 1 s). ROI, region of interest.
Figure 9
Example of long-term gene expression following hydrodynamic delivery into mice. Results of delivery of the human clotting Factor IX (hFIX) gene in a Sleeping Beauty (SB) transposon into FIX-deficient mice. Long-term expression depended on the presence of a functional transposase (SB10 regulated by a cytomegalovirus (CMV) promoter). The top portion of the figure shows the constructs delivered in cis. The graphs show the levels of hFIX in the mice as a function of time after hydrodynamic injection relative to normal enzyme amounts (therapeutic level). This figure is adapted from Figure 4 of Yant et al. (2000).
Similar articles
- A Transposon-Based Mouse Model of Hepatocellular Carcinoma via Hydrodynamic Tail Vein Injection.
Yu S, Vernia S. Yu S, et al. Methods Mol Biol. 2020;2164:129-143. doi: 10.1007/978-1-0716-0704-6_14. Methods Mol Biol. 2020. PMID: 32607890 - Lung-directed gene therapy in mice using the nonviral Sleeping Beauty transposon system.
Belur LR, Podetz-Pedersen K, Frandsen J, McIvor RS. Belur LR, et al. Nat Protoc. 2007;2(12):3146-52. doi: 10.1038/nprot.2007.460. Nat Protoc. 2007. PMID: 18079714 - Gene expression in lung and liver after intravenous infusion of polyethylenimine complexes of Sleeping Beauty transposons.
Podetz-Pedersen KM, Bell JB, Steele TW, Wilber A, Shier WT, Belur LR, McIvor RS, Hackett PB. Podetz-Pedersen KM, et al. Hum Gene Ther. 2010 Feb;21(2):210-20. doi: 10.1089/hum.2009.128. Hum Gene Ther. 2010. PMID: 19761403 Free PMC article. - Sleeping beauty transposon-mediated gene therapy for prolonged expression.
Hackett PB, Ekker SC, Largaespada DA, McIvor RS. Hackett PB, et al. Adv Genet. 2005;54:189-232. doi: 10.1016/S0065-2660(05)54009-4. Adv Genet. 2005. PMID: 16096013 Review. - Efficacy and safety of Sleeping Beauty transposon-mediated gene transfer in preclinical animal studies.
Hackett PB Jr, Aronovich EL, Hunter D, Urness M, Bell JB, Kass SJ, Cooper LJ, McIvor S. Hackett PB Jr, et al. Curr Gene Ther. 2011 Oct;11(5):341-9. doi: 10.2174/156652311797415827. Curr Gene Ther. 2011. PMID: 21888621 Free PMC article. Review.
Cited by
- Sleeping Beauty mRNA-LNP enables stable rAAV transgene expression in mouse and NHP hepatocytes and improves vector potency.
Zakas PM, Cunningham SC, Doherty A, van Dijk EB, Ibraheim R, Yu S, Mekonnen BD, Lang B, English EJ, Sun G, Duncan MC, Benczkowski MS, Altshuler RC, Singh MJ, Kibbler ES, Tonga GY, Wang ZJ, Wang ZJ, Li G, An D, Rottman JB, Bhavsar Y, Purcell C, Jain R, Alberry R, Roquet N, Fu Y, Citorik RJ, Rubens JR, Holmes MC, Cotta-Ramusino C, Querbes W, Alexander IE, Salomon WE. Zakas PM, et al. Mol Ther. 2024 Oct 2;32(10):3356-3371. doi: 10.1016/j.ymthe.2024.06.021. Epub 2024 Jul 8. Mol Ther. 2024. PMID: 38981468 - HDI-STARR-seq: Condition-specific enhancer discovery in mouse liver in vivo.
Chang TY, Waxman DJ. Chang TY, et al. bioRxiv [Preprint]. 2024 Jun 12:2024.06.10.598329. doi: 10.1101/2024.06.10.598329. bioRxiv. 2024. PMID: 38915578 Free PMC article. Preprint. - Polymer design via SHAP and Bayesian machine learning optimizes pDNA and CRISPR ribonucleoprotein delivery.
Dalal RJ, Oviedo F, Leyden MC, Reineke TM. Dalal RJ, et al. Chem Sci. 2024 Apr 22;15(19):7219-7228. doi: 10.1039/d3sc06920f. eCollection 2024 May 15. Chem Sci. 2024. PMID: 38756796 Free PMC article. - Hepatic IRE1α-XBP1 signaling promotes GDF15-mediated anorexia and body weight loss in chemotherapy.
Tang Y, Yao T, Tian X, Xia X, Huang X, Qin Z, Shen Z, Zhao L, Zhao Y, Diao B, Ping Y, Zheng X, Xu Y, Chen H, Qian T, Ma T, Zhou B, Xu S, Zhou Q, Liu Y, Shao M, Chen W, Shan B, Wu Y. Tang Y, et al. J Exp Med. 2024 Jul 1;221(7):e20231395. doi: 10.1084/jem.20231395. Epub 2024 May 2. J Exp Med. 2024. PMID: 38695876 Free PMC article. - Cancer cell genetics shaping of the tumor microenvironment reveals myeloid cell-centric exploitable vulnerabilities in hepatocellular carcinoma.
Ramirez CFA, Taranto D, Ando-Kuri M, de Groot MHP, Tsouri E, Huang Z, de Groot D, Kluin RJC, Kloosterman DJ, Verheij J, Xu J, Vegna S, Akkari L. Ramirez CFA, et al. Nat Commun. 2024 Mar 22;15(1):2581. doi: 10.1038/s41467-024-46835-2. Nat Commun. 2024. PMID: 38519484 Free PMC article.
References
- Muruve DA, Barnes MJ, Stillman IE, Libermann TA. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Hum. Gene Ther. 1999;10:965–976. - PubMed
- Schroder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. - PubMed
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. - PubMed
- Nakai H, et al. AAV serotype 2 vectors preferentially integrate into active genes in mice. Nat. Genet. 2003;34:297–302. - PubMed
MeSH terms
Substances
Grants and funding
- R43 HL072539-01/HL/NHLBI NIH HHS/United States
- P01 HD032652-120006/HD/NICHD NIH HHS/United States
- P01 HD032652-100006/HD/NICHD NIH HHS/United States
- P01 HD032652/HD/NICHD NIH HHS/United States
- P01 HD032652-09A10006/HD/NICHD NIH HHS/United States
- P01 HD032652-110006/HD/NICHD NIH HHS/United States
- R43 HL072539/HL/NHLBI NIH HHS/United States
- P01 HD032652-130006/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical