Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila - PubMed (original) (raw)
Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila
Tatiana I Gerasimova et al. Mol Cell. 2007.
Abstract
CTCF plays a central role in vertebrate insulators and forms part of the Fab-8 insulator in Drosophila. dCTCF is present at hundreds of sites in the Drosophila genome, where it is located at the boundaries between bands and interbands in polytene chromosomes. dCTCF colocalizes with CP190, which is required for proper binding of dCTCF to chromatin, but not with the other gypsy insulator proteins Su(Hw) or Mod(mdg4)2.2. Mutations in the CP190 gene affect Fab-8 insulator activity, suggesting that CP190 is an essential component of both gypsy and dCTCF insulators. dCTCF is present at specific nuclear locations, forming large insulator bodies that overlap with those formed by Su(Hw), Mod(mdg4)2.2, and CP190. The results suggest that Su(Hw) and dCTCF may be the DNA-binding components of two different subsets of insulators that share CP190 and cooperate in the formation of insulator bodies to regulate the organization of the chromatin fiber in the nucleus.
Figures
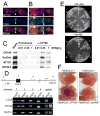
Figure 1. Mutations in the dCTCF gene affect Fab-8 insulator activity
(A) Protein extracts from larvae of various fly strains were subjected to Western analysis. Lane 1, wild type OreR; lane 2, P{EPgy2}CTCFEY15833; lane 3, dCTCFy+1; lane 4, dCTCFy+2; lane 5, dCTCFy+6. (B) Eyes of male and female flies of the genotypes Fab-860.39.2/CyO; dCTCFy+6 (top) and _Fab-_860.39.2/CyO; +/TM6 (bottom). (C) Abdomens of pharate adult flies pulled from the pupal cases. The left panel shows the abdomen of a wild type fly whereas the right panel shows the abdomen of a fly homozygous for the dCTCFy+6 allele; the arrow points to an additional abdominal segment posterior to A6.
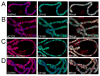
Figure 2. Distribution of dCTCF on polytene chromosomes of wild type third instar larvae in relation to DNA and gypsy insulator proteins. DNA is stained with DAPI (blue) in all immunofluorescence images
Immunolocalization of (A) dCTCF (red) on polytene chromosomes. Arrows point to the location of dCTCF sites at the boundary of DAPI band and interbands. (B) dCTCF (red) and Su(Hw) (green) on polytene chromosomes. (C) dCTCF (green) and Mod(mdg4)2.2 (red) on polytene chromosomes. (D) dCTCF (red) and CP190 (green) on polytene chromosomes.
Figure 3. Interaction of dCTCF with gypsy insulator proteins
(A) dCTCF is present at the cytological location of the Fab-8 insulator. Red indicates the immunolocalization of dCTCF, green indicates the localization of Fab-8 sequences by FISH and DNA is stained with DAPI in blue. Arrow points to the location of Fab-8. (B) CP190 is present at the cytological location of the Fab-8 insulator. Red indicates the immunolocalization of CP190, green indicates the localization of Fab-8 sequences by FISH and DNA is stained with DAPI in blue. Arrow points to the location of Fab-8. (C) Western analysis of control preimmune (lanes 2–4) and α-CP190 column (lanes 5–7) immunoaffinity purifications to verify the presence of CP190, Su(Hw), dCTCF and Mod(mdg4)2.2 in α-CP190 eluates at the indicated concentrations of MgCl2. (D) Chromatin immunoprecipitation analysis of the Fab-8 insulator. The top part of the panel is a diagram of the 5′ region of the Abd-B gene, including the Fab-8 insulator. Numbered arrows above the diagram indicate the location of primers used in the PCR reactions. The lower portion of the panel shows the result of the electropheretic analysis of the PCR products after immunoprecipitation with antibodies to the proteins indicated on the left. For each primer pair, an input and IP lane are shown. Also shown is a control using primers to the insulator present in the gypsy retrotransposon. (E) Growth of yeast strains expressing dCTCF and Su(Hw), CP190 or control on nonselective (+histidine +adenine) (top) or selective ( histidine -adenine) (bottom) media. Sector 1, Su(Hw)AD-CP190BD. Sector 2, dCTCFAD-CP190BD. Sector3, dCTCFAD-CP190-BTB domainBD. Sector 4, dCTCFAD-CP190-carboxytermBD. Sector 5, dCTCFAD-emptyBD. Sector 6, CP190BD-emptyAD. (F) Left panel: Eyes of male flies of the genotype Fab-860.39.2/CyO;+/+ (top) and Fab-860.39.2/CyO; CP1904-1/CP190H31-2 (bottom). Right panel: Eyes of male flies of the genotype _Fab-_860.39.2/CyO;+/+ (top) and Fab-860.39.2/CyO; su(Hw)2 (bottom).
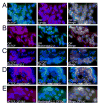
Figure 4. Levels of dCTCF in wild type and mutant flies. DNA is stained with DAPI (blue) in all panels
(A) Top row. Immunolocalization of CP190 (red) and of Lip (control, green) in polytene chromosomes from wild type OreR larvae. Bottom row. Immunolocalization of CP190 (red) and of Lip (control, green) in polytene chromosomes from dCTCFy+6/dCTCFy+6 mutant larvae. (B) Top row. Immunolocalization of dCTCF (red) and of Lip (control, green) in polytene chromosomes from wild type larvae. Bottom row. Immunolocalization of dCTCF (red) and of Lip (control, green) in polytene chromosomes from CP190P1/CP190P11 mutant larvae. (C) Western analysis of protein extracts from wild type (OR) and CP190P1/CP190P11 larvae. Lamin was used as a control. (D) Western analysis of protein extracts from wild type (OR) and dCTCFy+6/dCTCFy+6 larvae. Lamin was used as a control.
Figure 5. Co-localization of dCTCF and gypsy insulator proteins on polytene chromosomes from wild type larvae. DNA is stained with DAPI (blue) in all panels
(A) Su(Hw) (red) and CP190 (green) on polytene chromosomes. (B) dCTCF (red) and CP190 (green) on polytene chromosomes. (C) dCTCF plus Su(Hw) (red) and CP190 (green) on polytene chromosomes. (D) Su(Hw) (red) and CP190 (green) on polytene chromosomes of dCTCFy+6 mutants.
Figure 6. Localization of dCTCF and gypsy insulator proteins in diploid nuclei of wild type and mutant larvae. DNA is stained with DAPI (blue) in all images
(A) Immunolocalization of dCTCF (green) and CP190 (red) in diploid nuclei from imaginal disc cells of wild type larvae. (B) Immunolocalization of dCTCF (red) and Mod(mdg4)2.2 (green) in diploid nuclei from imaginal disc cells of wild type larvae. (C) Immunolocalization of Mod(mdg4)2.2 (red) and CP190 (green) in diploid nuclei from imaginal disc cells of dCTCFy+6/dCTCFy+6 mutant larvae. (D) Immunolocalization of dCTCF (red) and CP190 (green) in diploid nuclei from imaginal disc cells of mod(mdg4)T6 mutant larvae. (E) Immunolocalization of dCTCF (red) and Mod(mdg4)2.2 (green) in diploid nuclei from imaginal disc cells of CP190P1/CP190P11 mutant larvae.
Figure 7. Possible models to explain the formation of insulator bodies by dCTCF. In both panels insulator proteins are represented by differently colored ovals: Su(Hw) is shown in blue, Mod(mdg4)2.2 in green, dCTCF in purple and CP190 in pink
(A) Su(Hw) and dCTCF bind different insulator sequences but coalesce in the same insulator bodies through interactions between the CP190 protein present in both insulators (B) Su(Hw) and dCTCF form independent insulator bodies but both co-localize in the same region of the nucleus. This may imply the existence of special nuclear compartments where insulator bodies are located.
Similar articles
- EAST Organizes Drosophila Insulator Proteins in the Interchromosomal Nuclear Compartment and Modulates CP190 Binding to Chromatin.
Golovnin A, Melnikova L, Shapovalov I, Kostyuchenko M, Georgiev P. Golovnin A, et al. PLoS One. 2015 Oct 21;10(10):e0140991. doi: 10.1371/journal.pone.0140991. eCollection 2015. PLoS One. 2015. PMID: 26489095 Free PMC article. - The centrosomal protein CP190 is a component of the gypsy chromatin insulator.
Pai CY, Lei EP, Ghosh D, Corces VG. Pai CY, et al. Mol Cell. 2004 Dec 3;16(5):737-48. doi: 10.1016/j.molcel.2004.11.004. Mol Cell. 2004. PMID: 15574329 - The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning.
Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, Renkawitz R. Mohan M, et al. EMBO J. 2007 Oct 3;26(19):4203-14. doi: 10.1038/sj.emboj.7601851. Epub 2007 Sep 6. EMBO J. 2007. PMID: 17805343 Free PMC article. - Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.
Ahanger SH, Shouche YS, Mishra RK. Ahanger SH, et al. Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review. - Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila.
Gdula DA, Gerasimova TI, Corces VG. Gdula DA, et al. Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9378-83. doi: 10.1073/pnas.93.18.9378. Proc Natl Acad Sci U S A. 1996. PMID: 8790337 Free PMC article. Review.
Cited by
- Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks.
Chetverina D, Aoki T, Erokhin M, Georgiev P, Schedl P. Chetverina D, et al. Bioessays. 2014 Feb;36(2):163-72. doi: 10.1002/bies.201300125. Epub 2013 Nov 26. Bioessays. 2014. PMID: 24277632 Free PMC article. Review. - DNA topoisomerase II modulates insulator function in Drosophila.
Ramos E, Torre EA, Bushey AM, Gurudatta BV, Corces VG. Ramos E, et al. PLoS One. 2011 Jan 27;6(1):e16562. doi: 10.1371/journal.pone.0016562. PLoS One. 2011. PMID: 21304601 Free PMC article. - EAST affects the activity of Su(Hw) insulators by two different mechanisms in Drosophila melanogaster.
Melnikova L, Shapovalov I, Kostyuchenko M, Georgiev P, Golovnin A. Melnikova L, et al. Chromosoma. 2017 Mar;126(2):299-311. doi: 10.1007/s00412-016-0596-3. Epub 2016 Apr 30. Chromosoma. 2017. PMID: 27136940 - Conserved boundary elements from the Hox complex of mosquito, Anopheles gambiae.
Ahanger SH, Srinivasan A, Vasanthi D, Shouche YS, Mishra RK. Ahanger SH, et al. Nucleic Acids Res. 2013 Jan;41(2):804-16. doi: 10.1093/nar/gks1178. Epub 2012 Dec 4. Nucleic Acids Res. 2013. PMID: 23221647 Free PMC article. - Chromatin insulators: linking genome organization to cellular function.
Phillips-Cremins JE, Corces VG. Phillips-Cremins JE, et al. Mol Cell. 2013 May 23;50(4):461-74. doi: 10.1016/j.molcel.2013.04.018. Mol Cell. 2013. PMID: 23706817 Free PMC article. Review.
References
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab- 7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. - PubMed
- Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. - PubMed
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous