Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila - PubMed (original) (raw)
Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila
Deborah L Berry et al. Cell. 2007.
Abstract
Autophagy is a catabolic process that is negatively regulated by growth and has been implicated in cell death. We find that autophagy is induced following growth arrest and precedes developmental autophagic cell death of Drosophila salivary glands. Maintaining growth by expression of either activated Ras or positive regulators of the class I phosphoinositide 3-kinase (PI3K) pathway inhibits autophagy and blocks salivary gland cell degradation. Developmental degradation of salivary glands is also inhibited in autophagy gene (atg) mutants. Caspases are active in PI3K-expressing and atg mutant salivary glands, and combined inhibition of both autophagy and caspases increases suppression of gland degradation. Further, induction of autophagy is sufficient to induce premature cell death in a caspase-independent manner. Our results provide in vivo evidence that growth arrest, autophagy, and atg genes are required for physiological autophagic cell death and that multiple degradation pathways cooperate in the efficient clearance of cells during development.
Figures
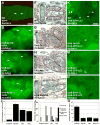
Figure 1. Loss of cortical tGPH coincides with growth arrest and the induction of autophagy
(A–C) Cortical localization of tGPH reflects PI3K activity during normal development of fkhGAL4; tGPH pupae. (A) Cortical tGPH is observed in the growing salivary glands of wandering larvae. (B) No cortical tGPH is observed in salivary glands 6 hours apf or (C) in dying 13.5-hour salivary glands. (D–G) Autophagy is indicated by GFP spots (autophagosomes) in salivary glands during normal development of fkhGAL4; UAS-GFP-LC3 pupae. (D) GFP-LC3 spots are rare in the growing salivary glands of feeding larvae. (E) GFP-LC3 spots were detected at low levels 6 hours apf, and were abundant in salivary glands 13.5 hours apf (F). (G) The number of GFP-LC3 spots per image was quantified in salivary glands of feeding larvae and of pupae 6 and 13.5 hours apf, and are represented as the mean ± SE. Scale bar in (A) is 100 μm, and (A–C) are same magnification. Arrows point to cell cortex in (A–C). Scale bar in (D) is 25 μm, and (D–F) are same magnification. Arrows point to GFP-LC3 spots in (D–F).
Figure 2. Maintenance of growth prevents induction of autophagy and inhibits salivary gland degradation
(A–D) Cortical localization of tGPH correlates with growth. (A) At 13.5 hours apf, no cortical tGPH is observed in control dying salivary glands. Expression of either (B) Dp110, (C) Akt or (D) RasV12 in the salivary glands results in increased growth and maintenance of cortically localized tGPH 13.5 hours apf. (E) Cell area measurements in the indicated genotypes 13.5 hours apf illustrate the increase in cell size in Dp110, Akt, and RasV12 expressing salivary glands. The data are presented as fold increase in cell area compared to cells from control fkhGAL4; Canton-S pupae and are represented as mean ± SE. (F–I) Paraffin sections of pupae 24 hours apf. (F) Salivary glands are completely degraded in control pupae (Canton-S/UAS-Dp110) 24 hours apf. The location where glands resided prior to degradation is encircled. Maintaining growth by expression of either (G) Dp110 (H), Akt, or (I) RasV12 in salivary glands inhibits degradation and tissue is present 24 hours apf. Circles outline the persistent salivary gland tissue in these pupae. (J) Paraffin sections of the above genotypes were evaluated for the amount and type of salivary gland tissue that was present in 24 hour pupae. The percentage of pupae with each phenotype is presented (n > 20 pupae/genotype). (K–N) GFP-LC3 spots are abundant in control salivary glands 13.5 hours apf (K), but are reduced in 13.5 hour salivary glands expressing (L) Dp110, (M) Akt, and (N) RasV12. (O) The number of GFP-LC3 spots per image was quantified for the indicated genotypes 13.5 hours apf. The data is represented as mean ± SE. Scale bar in (A) is 60 μm and (A–D) are the same magnification. Scale bar in (F) is 200 μm and (F–I) are the same magnification. Scale bar in (K) is 19 μm and (K–N) are the same magnification. Arrows point to the cell cortex in (A–D). Arrows point to GFP-LC3 spots in (K–N). Symbols are (b) brain and (g) gut.
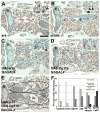
Figure 3. Ras and PI3K inhibition of salivary gland degradation requires TOR
Paraffin sections 24 hours apf show that increasing the cell cycle by expression of either (A) Myc or (B) CycD with Cdk4 in the salivary gland does not prevent salivary gland degradation. Inhibition of salivary gland degradation by expression of (C) RasV12 in the salivary glands (circled) is ameliorated by co-expression of (D) RasV12 with TORted. Similarly, inhibition of salivary gland degradation by expression of (E) Dp110 in the salivary glands is ameliorated by co-expression of (F) Dp110 with TORted. Scale bar in (A) is 200 μm and (A–F) are the same magnification. Red boxes indicate the region of higher magnification shown in the insets for each image. Red arrows point to salivary gland fragments. Black arrowheads point to fat body that is present in the area after the salivary gland has degraded. Symbols are (b) brain, (m) muscle and (g) gut.
Figure 4. DNA is fragmented and caspases are active when salivary gland growth is maintained
(A–F) Visualization of DNA fragmentation by the TUNEL assay. Nuclei of 6-hour apf (A) Canton-S control, (B) Dp110-, and (C) RasV12-expressing salivary glands are TUNEL negative (black triangles) while nuclei in nearby dying gut cells are TUNEL positive (black arrows). TUNEL positive nuclei were detected in salivary glands from 13.5-hour (D) Canton-S control, (E) Dp110- and (F) RasV12-expressing salivary glands. (G, H) Antibodies against processed caspase-3 (green) and nuclear lamins (red) indicate that processed caspase-3 is present (white arrows) and that the caspase substrate lamin is degrading (white arrowheads) in 13.5-hour (G) wild-type control glands and (H) Dp110-expressing glands (DNA is blue). (I) Cleavage of the caspase substrate Z-DEVD-AMC was measured in 13.5-hour salivary glands of Control Canton-S, Control Canton-S plus Ac-DEVD-CHO as an inhibitor, and of p35-, Dp110-, Akt- and RasV12-expressing salivary glands. Scale bar in (A) is 50 μm and (A–F) are the same magnification. Scale bar in (G) is 30 μm and (G–H) are the same magnification. Symbols are (sg) salivary gland, (g) gut, and (RFU) relative fluorescence units.
Figure 5. Multiple degradation pathways are required for complete salivary gland histolysis
(A–E) Paraffin sections 24 hours apf. Salivary glands are degraded in (A) arkL46/N28 and (B) droncI24/I29 mutants that develop normally. Partial degradation occurs in salivary glands expressing (C) p35 and (D) Dp110. (E) Expression of p35 combined with Dp110 significantly increases the amount of persistent salivary gland tissue (encircled). (F) Histological sections of the indicated genotypes were evaluated for the amount and type of salivary gland tissue that was present in pupae 24 hours apf. The percentage of pupae with each phenotype is presented (n > 20 pupae/genotype). Scale bar in (A) is 200 μm and (A–E) are the same magnification. Red boxes indicate the region of higher magnification shown in the insets for each image. Red arrows point to salivary gland fragments. Black arrowheads point to fat body that is present in the area after the salivary gland has degraded. Symbols are (b) brain, (g) gut, (m) muscle, and (lu) lumen.
Figure 6. Loss-of-function autophagy mutants inhibit salivary gland cell degradation in the presence of caspases
(A–E) Histological sections 24 hours apf. (A) Salivary glands are degraded in atg8aKG07569/Canton-S control pupae. (B) atg8aKG07569 mutant pupae contain vacuolated salivary gland cellular fragments. (C) Heat shock induced expression of hsdAtg8a-GFP rescued the vacuolated salivary gland phenotype of atg8aKG07569 mutant pupae. (D) Atg1KQ expression prevents complete salivary gland destruction and vacuolated salivary gland cell fragments persist. (E) atg18KG03090/Df(3L)66C-G28 mutant pupae contain vacuolated salivary gland cell fragments. (F) Cleavage of the caspase substrate DEVD-AMC was measured in whole pupae at 4 hours apf in: Canton-S control, Canton-S plus Ac-DEVD-CHO inhibitor, atg8aKG07569 mutant pupae and atg18KG03090/Df(3L)Exel6112 mutant pupae. Data is presented as the mean ± SE. (G–H) TUNEL positive nuclei (black arrows) are present in salivary glands of (G) atg8aKG07569 and (H) atg18KG03090/Df(3L)Exel6112 mutant pupae. (I) Expression of p35 in the salivary glands of atg18KG03090/Df(3L)Exel6112 mutant pupae leads to increased salivary gland persistence. Scale bar in (A) is 200 μm and (A–E and I) are the same magnification. Scale bar in (G) is 50 μm and (G–H) are the same magnification. Red boxes indicate the region of higher magnification shown in the insets for each image. Red arrows point to salivary gland fragments. Black arrowheads point to fat body that is present in the area after the salivary gland has degraded. Symbols are (b) brain, (g) gut and (lu) lumen.
Figure 7. Expression of Atg1 induces caspase-independent degradation of salivary glands
(A–B) Lysotracker staining detects autophagy. (A) Little to no lysotracker staining is observed in Canton-S; fkhGAL4 control salivary glands from feeding larvae. (B) Many lysotracker positive spots are detected in feeding larval salivary glands expressing Atg1GS10797 (white arrows). (C) Paraffin sections 6 hours apf show that intact glands are present in UAS-Atg1/Canton-S control pupae. (D) Atg1GS10797 expression causes premature degradation of salivary glands. (E) No TUNEL-positive nuclei are present 6 hours apf in Atg1GS10797-expressing salivary glands. Black arrowheads point to TUNEL-negative nuclei within the salivary glands. Black arrows indicate TUNEL-positive cells in adjacent tissues. (F) Inhibition of caspases by co-expression of p35 does not suppress the Atg1GS10797 cell degradation phenotype. (G) Inhibition of autophagy by co-expression of atg12-IR suppresses the Atg1GS10797 cell degradation phenotype. (H) Co-expression of Atg1GS10797 with Dp110 leads to complete salivary gland degradation. Scale bar in (A) is 75 μm and (A–B) are the same magnification. Scale bar in (C) is 200 μm and (C–D and F–H) are the same magnification. Scale bar in (E) is 50 μm. Red boxes indicate the region of higher magnification shown in the insets for each image. Symbols are (b) brain, (sg) salivary gland, and (g) gut.
Comment in
- Autophagy and cell death: no longer at odds.
Bergmann A. Bergmann A. Cell. 2007 Dec 14;131(6):1032-4. doi: 10.1016/j.cell.2007.11.027. Cell. 2007. PMID: 18083090 Free PMC article.
Similar articles
- Warts is required for PI3K-regulated growth arrest, autophagy, and autophagic cell death in Drosophila.
Dutta S, Baehrecke EH. Dutta S, et al. Curr Biol. 2008 Oct 14;18(19):1466-75. doi: 10.1016/j.cub.2008.08.052. Epub 2008 Sep 25. Curr Biol. 2008. PMID: 18818081 Free PMC article. - Activation of autophagy during cell death requires the engulfment receptor Draper.
McPhee CK, Logan MA, Freeman MR, Baehrecke EH. McPhee CK, et al. Nature. 2010 Jun 24;465(7301):1093-6. doi: 10.1038/nature09127. Nature. 2010. PMID: 20577216 Free PMC article. - Dynein light chain 1 is required for autophagy, protein clearance, and cell death in Drosophila.
Batlevi Y, Martin DN, Pandey UB, Simon CR, Powers CM, Taylor JP, Baehrecke EH. Batlevi Y, et al. Proc Natl Acad Sci U S A. 2010 Jan 12;107(2):742-7. doi: 10.1073/pnas.0907967107. Epub 2009 Dec 22. Proc Natl Acad Sci U S A. 2010. PMID: 20080745 Free PMC article. - Autophagic programmed cell death in Drosophila.
Baehrecke EH. Baehrecke EH. Cell Death Differ. 2003 Sep;10(9):940-5. doi: 10.1038/sj.cdd.4401280. Cell Death Differ. 2003. PMID: 12934068 Review. - Salivary cellular signaling and gene regulation.
Wen X, Lin HH, Ann DK. Wen X, et al. Adv Dent Res. 2000 Dec;14:76-80. doi: 10.1177/08959374000140011201. Adv Dent Res. 2000. PMID: 11842928 Review.
Cited by
- The Drosophila TIPE family member Sigmar interacts with the Ste20-like kinase Misshapen and modulates JNK signaling, cytoskeletal remodeling and autophagy.
Chittaranjan S, Xu J, Kuzyk M, Dullat HK, Wilton J, DeVorkin L, Lebovitz C, Morin GB, Marra MA, Gorski SM. Chittaranjan S, et al. Biol Open. 2015 Apr 2;4(5):672-84. doi: 10.1242/bio.20148417. Biol Open. 2015. PMID: 25836674 Free PMC article. - Uba1 functions in Atg7- and Atg3-independent autophagy.
Chang TK, Shravage BV, Hayes SD, Powers CM, Simin RT, Wade Harper J, Baehrecke EH. Chang TK, et al. Nat Cell Biol. 2013 Sep;15(9):1067-78. doi: 10.1038/ncb2804. Epub 2013 Jul 21. Nat Cell Biol. 2013. PMID: 23873149 Free PMC article. - The role of autophagy in Drosophila metamorphosis.
Tracy K, Baehrecke EH. Tracy K, et al. Curr Top Dev Biol. 2013;103:101-25. doi: 10.1016/B978-0-12-385979-2.00004-6. Curr Top Dev Biol. 2013. PMID: 23347517 Free PMC article. Review. - The Role of Mosquito Hemocytes in Viral Infections.
Cardoso-Jaime V, Tikhe CV, Dong S, Dimopoulos G. Cardoso-Jaime V, et al. Viruses. 2022 Sep 20;14(10):2088. doi: 10.3390/v14102088. Viruses. 2022. PMID: 36298644 Free PMC article. Review. - Autophagy drives epidermal deterioration in a Drosophila model of tissue aging.
Scherfer C, Han VC, Wang Y, Anderson AE, Galko MJ. Scherfer C, et al. Aging (Albany NY). 2013 Apr;5(4):276-87. doi: 10.18632/aging.100549. Aging (Albany NY). 2013. PMID: 23599123 Free PMC article.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11:36–42.
- Arsham AM, Neufeld TP. Thinking globally and acting locally with TOR. Curr Opin Cell Biol. 2006;18:589–597. - PubMed
- Baehrecke EH. How death shapes life during development. Nature Reviews Mol Cell Biol. 2002;3:779–787. - PubMed
- Baehrecke EH. Autophagy: dual roles in life and death? Nature Reviews Mol Cell Biol. 2005;6:505–510. - PubMed
- Britton JS, Lockwood WK, Li L, Cohen SM, Edgar BA. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev Cell. 2002;2:239–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM059136-07/GM/NIGMS NIH HHS/United States
- R01 GM059136-06/GM/NIGMS NIH HHS/United States
- R01 GM079431-01/GM/NIGMS NIH HHS/United States
- R01 GM059136/GM/NIGMS NIH HHS/United States
- R56 GM059136/GM/NIGMS NIH HHS/United States
- R01 GM079431/GM/NIGMS NIH HHS/United States
- R01 GM059136-08/GM/NIGMS NIH HHS/United States
- GM079431/GM/NIGMS NIH HHS/United States
- R01 GM059136-09/GM/NIGMS NIH HHS/United States
- GM59136/GM/NIGMS NIH HHS/United States
- R01 GM059136-10/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases