cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans - PubMed (original) (raw)
cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans
Craig T Miller et al. Cell. 2007.
Abstract
Dramatic pigmentation changes have evolved within most vertebrate groups, including fish and humans. Here we use genetic crosses in sticklebacks to investigate the parallel origin of pigmentation changes in natural populations. High-resolution mapping and expression experiments show that light gills and light ventrums map to a divergent regulatory allele of the Kit ligand (Kitlg) gene. The divergent allele reduces expression in gill and skin tissue and is shared by multiple derived freshwater populations with reduced pigmentation. In humans, Europeans and East Asians also share derived alleles at the KITLG locus. Strong signatures of selection map to regulatory regions surrounding the gene, and admixture mapping shows that the KITLG genomic region has a significant effect on human skin color. These experiments suggest that regulatory changes in Kitlg contribute to natural variation in vertebrate pigmentation, and that similar genetic mechanisms may underlie rapid evolutionary change in fish and humans.
Figures
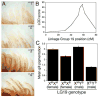
Figure 1. A QTL with a major effect on gill pigmentation
(A) Segregating variation in melanocyte distribution within the gills of Japanese marine × Paxton benthic F2 fish. F2 fish show a range of gill pigmentation, from gills with few melanocytes and overall white appearance (class “1”), to intermediate (classes “2” and “3”), to gills with many melanocytes and overall dark brownish appearance (class “4”). (B) Mapping results on linkage group 19. Genetic distance in centiMorgans (cM) is on the x-axis, and LOD score on the y-axis. Significance threshold is 4.5 (van Ooijen, 1999). Markers from left to right are Stn303, Stn185, Stn186, Stn187, Stn193, Stn194, Stn191, Stn398, Stn399, Stn400. (C) Gill pigmentation phenotypes of F2 fish according to genotype at Stn191, the peak QTL marker (mean scores +/− SEM). Since the grandparents were a Japanese marine female and Paxton benthic male, F2 fish can have any of four LG19 genotypes. Females can either be homozygous for marine chromosomes (XMXM) or heterozygous for marine and Paxton benthic chromosomes (XMXB). Males all have the benthic Y chromosome (YB), and can have a marine or freshwater X chromosome. Differences between genotypic classes are highly significant for both males (p = 9 × 10−40) and females (p = 2 × 10−8).
Figure 2. Fine mapping the LG19 pigmentation QTL
(A) X chromosome genotypes and gill pigmentation phenotypes of recombinant F2 males. Each row is an informative recombinant F2 male. Light (yellow) or dark (gray) gill score is shown in the first column, followed by genotype at markers along the X chromosome. Positions at top correspond to location in megabases of scaffold 3 from the stickleback genome assembly (Broad Institute, 2006). The first and last marker columns are Stn194 and Stn398, the markers flanking the QTL peak in Fig 1B. Microsatellites are listed in Table S1. Genotypes are coded yellow for Paxton benthic, gray for marine. The 315 kb minimal region concordant with gill pigmentation phenotype is boxed in blue. (B) Schematic of the 315 kb QTL interval, which contains 15 genes including Kit ligand (Kitlg). Genes are listed in Table S2.
Figure 3. LG19 pigmentation QTL controls melanization of ventral skin
(A) Schematic of region shown in B and C (box). The red dashed line between the posterior lateral line and the cloaca shows where melanization was scored in (D). (B) F2 male with marine X chromosome and heavily melanized ventrum. A nearly continuous field of melanocytes covers the ventrum. (C) F2 male with Paxton benthic X chromosome and sparsely melanized ventrum. (D) Mean percentage of ventral flank melanization for F2s of each possible LG19 QTL genotype. Each genotypic class contains from 24 to 31 F2 fish non-recombinant for the _Stn194_-Stn398 interval. Shown are the means +/− the SEM. Differences between classes are highly significant (p = 1 × 10−7 for one-way ANOVA) and LG19 genotype controls flank melanization in both males and females (XMYB vs. XBYB males, p = 4 × 10−9; XMXM vs. XMXB females, p = 0.017).
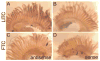
Figure 4. The freshwater Kit ligand variant is found in other populations with sparsely melanized gills and skin
(A) Map of west coast of North America showing locations of marine fish (Little Campbell River, BC; LITC) and derived freshwater populations (Paxton benthic, PAXB; Paxton limnetic, PAXL; Fishtrap Creek, FTC; and G. williamsoni, WMSO). (B) Neighbor-joining phylogenetic tree showing that FTC and WMSO have PAXB-like Kitlg alleles. In contrast, PAXL, which occurs as a species pair with PAXB, has marine-like alleles. (C) Nucleotide alignment of part of exon 8 and intron 8, showing that PAXB, FTC, and WMSO share a closely related Kitlg haplotype. (D) Ventrums (top) and gills (bottom) from populations shown in A-C. Ventrums are views like in Figure 3B,C. LITC and PAXL have heavily melanized ventrums and gills, while PAXB, FTC, and WMSO all have sparsely melanized ventrums and gills.
Figure 5. Reduced Kit ligand expression in gills of Fishtrap Creek fish
(A-D) In situ hybridization of gill arches from marine fish from Little Campbell River (LITC; A,B) and derived freshwater population Fishtrap Creek (FTC; C,D). (A,C) Antisense probe showing overall reduced Kitlg expression levels in FTC relative to LITC. (B,D) Sense probe controls showing staining observed is specific.
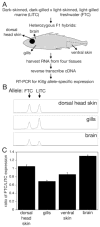
Figure 6. _cis_-regulatory changes in Kit ligand expression
(A) Schematic of allele-specific expression experiments. (B) Chromatogram showing representative results for size separation of Kitlg RT-PCR products from three different F1 hybrid tissues. Size is on the x-axis and fluorescence intensity on the y-axis. In dorsal head skin, near equal levels of each allele is amplified. In gills, the LITC allele is overexpressed relative to the FTC allele. In brains, the FTC allele is more abundant. (C) Ratio of freshwater to marine Kitlg expression in dorsal head skin, gills, ventral skin, and brain. Shown are means +/− SEM. For each tissue, at least eight F1 hybrids were analyzed. Expression in different tissues is significantly different (p = 1 × 10−14 by one-way ANOVA); as are all pairwise comparisons (p < 0.03 by Tuky’s HSD post hoc pairwise comparisons).
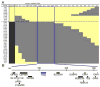
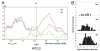
Figure 7. Effect of KITLG genotype on skin pigmentation in African-Americans
(A) Fay and Wu’s H from Haplotter (Voight et al., 2006) for a 1 Mb window centered on human KITLG. Strong signals of selection are detected 5′ (right) and 3′ (left) of KITLG in a population sample of European descent (CEU: CEPH, Utah residents with ancestry from northern and western Europe) and in a population sample from East Asia (ASN: Han Chinese in Beijing and Japanese in Tokyo). YRI: West Africans (Yoruba in Ibadan, Nigeria) (The International HapMap Consortium, 2005). A -log(Q) value of 3.0 indicates a value in the top one-thousandth of all scores in the human genome. (B) Histograms for melanin index (M) after adjustment for individual genetic ancestry in individuals with two European (GG), one European and one Afrrican (AG), or two African (AA) alleles at rs642742 upstream of the KITLG gene. Mean (SD) adjusted melanin indexes: for GG, −4.7 (7.7), n=18; for AG, −2.9 (8.8), n=121; for AA, 1.3 (9.1), n=232.
Comment in
- Sticklebacks and humans walk hand in fin to lighter skin.
Boughman JW. Boughman JW. Cell. 2007 Dec 14;131(6):1041-3. doi: 10.1016/j.cell.2007.11.029. Cell. 2007. PMID: 18083094
Similar articles
- Sticklebacks and humans walk hand in fin to lighter skin.
Boughman JW. Boughman JW. Cell. 2007 Dec 14;131(6):1041-3. doi: 10.1016/j.cell.2007.11.029. Cell. 2007. PMID: 18083094 - A pigment evolution Kitlg.
Greenhill ER, Kelsh RN. Greenhill ER, et al. Pigment Cell Melanoma Res. 2008 Apr;21(2):113-4. doi: 10.1111/j.1755-148X.2008.00447.x. Pigment Cell Melanoma Res. 2008. PMID: 18426403 No abstract available. - Predominance of _cis_-regulatory changes in parallel expression divergence of sticklebacks.
Verta JP, Jones FC. Verta JP, et al. Elife. 2019 May 15;8:e43785. doi: 10.7554/eLife.43785. Elife. 2019. PMID: 31090544 Free PMC article. - Fishing for the secrets of vertebrate evolution in threespine sticklebacks.
Peichel CL. Peichel CL. Dev Dyn. 2005 Dec;234(4):815-23. doi: 10.1002/dvdy.20564. Dev Dyn. 2005. PMID: 16252286 Review. - The genetic and molecular architecture of phenotypic diversity in sticklebacks.
Peichel CL, Marques DA. Peichel CL, et al. Philos Trans R Soc Lond B Biol Sci. 2017 Feb 5;372(1713):20150486. doi: 10.1098/rstb.2015.0486. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 27994127 Free PMC article. Review.
Cited by
- RNA sequencing reveals small RNAs differentially expressed between incipient Japanese threespine sticklebacks.
Kitano J, Yoshida K, Suzuki Y. Kitano J, et al. BMC Genomics. 2013 Apr 2;14:214. doi: 10.1186/1471-2164-14-214. BMC Genomics. 2013. PMID: 23547919 Free PMC article. - Genome Assembly Improvement and Mapping Convergently Evolved Skeletal Traits in Sticklebacks with Genotyping-by-Sequencing.
Glazer AM, Killingbeck EE, Mitros T, Rokhsar DS, Miller CT. Glazer AM, et al. G3 (Bethesda). 2015 Jun 3;5(7):1463-72. doi: 10.1534/g3.115.017905. G3 (Bethesda). 2015. PMID: 26044731 Free PMC article. - Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand.
Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G, Jiggins CD, Kronforst MR, Long AD, McMillan WO, Reed RD. Martin A, et al. Proc Natl Acad Sci U S A. 2012 Jul 31;109(31):12632-7. doi: 10.1073/pnas.1204800109. Epub 2012 Jul 16. Proc Natl Acad Sci U S A. 2012. PMID: 22802635 Free PMC article. - Extensive linkage disequilibrium and parallel adaptive divergence across threespine stickleback genomes.
Hohenlohe PA, Bassham S, Currey M, Cresko WA. Hohenlohe PA, et al. Philos Trans R Soc Lond B Biol Sci. 2012 Feb 5;367(1587):395-408. doi: 10.1098/rstb.2011.0245. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 22201169 Free PMC article. - Convergent evolution of RFX transcription factors and ciliary genes predated the origin of metazoans.
Chu JS, Baillie DL, Chen N. Chu JS, et al. BMC Evol Biol. 2010 May 4;10:130. doi: 10.1186/1471-2148-10-130. BMC Evol Biol. 2010. PMID: 20441589 Free PMC article.
References
- Bedell MA, Brannan CI, Evans EP, Copeland NG, Jenkins NA, Donovan PJ. DNA rearrangements located over 100 kb 5′ of the Steel (Sl)-coding region in Steel-panda and Steel-contrasted mice deregulate Sl expression and cause female sterility by disrupting ovarian follicle development. Genes Dev. 1995;9:455–470. - PubMed
- Bell MA, Foster SA. The Evolution of the Threespine Stickleback. Oxford University Press; Oxford: 1994.
- Bennett DC, Lamoreux ML. The color loci of mice--a genetic century. Pigment Cell Res. 2003;16:333–344. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases