ARS2 is a conserved eukaryotic gene essential for early mammalian development - PubMed (original) (raw)
ARS2 is a conserved eukaryotic gene essential for early mammalian development
Michael D Wilson et al. Mol Cell Biol. 2008 Mar.
Abstract
Determining the functions of novel genes implicated in cell survival is directly relevant to our understanding of mammalian development and carcinogenesis. ARS2 is an evolutionarily conserved gene that confers arsenite resistance on arsenite-sensitive Chinese hamster ovary cells. Little is known regarding the function of ARS2 in mammals. We report that ARS2 is transcribed throughout embryonic development and is expressed ubiquitously in mouse and human tissues. The mouse ARS2 protein is predominantly localized to the nucleus, and this nuclear localization is ablated in ARS2-null embryos, which in turn die around the time of implantation. After 24 h of culture, ARS2-null blastocysts contained a significantly greater number of apoptotic cells than wild-type or heterozygous blastocysts. By 48 h of in vitro culture, null blastocysts invariably collapsed and failed to proliferate. These data indicate ARS2 is essential for early mammalian development and is likely involved in an essential cellular process. The analysis of data from several independent protein-protein interaction studies in mammals, combined with functional studies of its Arabidopsis ortholog, SERRATE, suggests that this essential process is related to RNA metabolism.
Figures
FIG. 1.
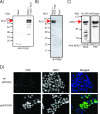
Overview and evolution of ARS2. (A) Phylogenetic analysis of the ARS2 genes from representative eukaryotic species. Amino acid alignments were made using CLUSTALW and manually edited. Columns containing gaps were deleted, and 224 informative sites were used to construct the phylogeny. A neighbor-joining tree using a Poisson distance matrix was made. (B) Alignment of the mouse ARS2 locus with representative mRNAs and genomic DNAs. Representative mRNAs and ESTs were prealigned by the University of California Santa Cruz using the BLAT algorithm. Genomic conservation of the ARS2 gene between mouse and seven additional species is shown as vertical black lines. The alignments were precomputed using the Multiz align algorithm (3) and can be found at positions chr5:136,243,845 to 136,260,875 of the mouse March 2005 assembly on the UCSC genome browser website.
FIG. 2.
Expression analysis of mouse and human ARS2 mRNAs. (A and B) Semiquantitative PCR analysis of normalized first-strand cDNAs in mouse and human tissues. The mouse 1,890-bp ARS2 product was observed in all tissues, and a mouse G3pdh primer set was used as a positive control. (C) Mouse ARS2 Northern blot. (Top) Normalized mouse Northern blot tissues probed with the ARS2 3′ probe. (Middle) Northern blot probed with the ARS2 5′ probe. (Bottom) Beta actin control probe. (D) (Top) Human ARS2 Northern blot probed with the ARS2 human 3′ probe. (Bottom) Northern blot probed with beta actin. (E) Normalized RNA blot of multiple human tissues using the ARS2 5′ cDNA as a probe.
FIG. 3.
ARS2 is expressed predominantly as a 100-kDa protein. (A to C) Western blot analysis of N terminus FLAG tag full-length mouse ARS2 protein stably transfected into the BOSC kidney cell line (A), C terminus MYC full-length mouse ARS2 fusion protein (transient transfection of BOSC cells) (B), and H1299 cell lysate using polyclonal and monoclonal anti-ARS2 antibodies raised against human ARS2 (C). (D) Anti-FLAG antibody staining of three-FLAG N-terminally labeled mouse ARS2 in a stably transfected BOSC cell line and a negative control in which the primary FLAG antibody was omitted (−ve primary). FITC, fluorescein isothiocyanate; DAPI, 4′,6′-diamidino-2-phenylindole.
FIG. 4.
Gene targeting of mouse ARS2. (A) Mouse ARS2 genomic locus and surrounding genes. Mouse exon positions, repetitive elements, neighboring genes, and thelocations of the arms of homology for gene targeting are shown to scale on the x axis. Percent identity shared with the orthologous human locus is shown on the y axis. The coordinates are based on the sequence AF312033. (B) Removal of the proximal promoter and exons 1 to 3 of mouse ARS2 using homologous recombination. Southern blot probes for BamHI and EcoRI digests are labeled P1 and P2, respectively. The targeting vector with a neomycin cassette (neo) and herpes simplex virus thymidine kinase (HSV-tk) are indicated, along with the arms of homology. (C) Southern blot screening of R1 ES cells: BamHI digestion and P1 produced a wild-type band of 9.6 kb and a mutant band of 5.6 kb, and EcoRI digestion and P2 produced a wild-type band of 14.7 kb and a mutant band of 12.0 kb. (D) Southern blot analysis of the ARS2 locus using BamHI and P1 after Cre-mediated removal of the neo cassette. (E) Southern blot analysis of E12.5 yolk sacs from two intercrosses of ARS2+/− mice using BamHI and P1. (F) PCR genotyping of blastocysts (E3.5) from ARS2+/− intercrosses. The wild-type band is 333 bp, and the mutant band is 563 bp.
FIG. 5.
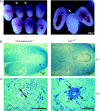
Dissections and histological analysis of wild-type and presumptive _ARS2_−/− embryos at E7.5 and E5.5. (A) Representative example of relatively small deciduas routinely obtained from ARS2+/− intercrosses at E7.5 (arrows). Embryos were never recovered from within these deciduas or small deciduas from E6.5. (B) Serial sectioning of random large and small deciduas at E7.5 revealed no discernible embryonic structure in presumptive mutants (n = 2). (C) Transverse, serial sectioning of E5.5 implantations from ARS2+/− intercrosses revealed a clear outer boundary of the outer zone of the decidual reaction (zd) in both presumptive _ARS2_−/− and ARS2+/? embryos. A small mass of abnormal cells with pycnotic nuclei was characteristic of presumptive _ARS2_−/− embryos (n = 3). Wild-type morphology containing a visceral endoderm (ve), parietal endoderm (pe), epiblast (epi), and proamniotic cavity was observed for all ARS2+/? embryos.
FIG. 6.
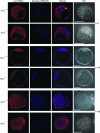
ARS2 distribution in ARS2+/+ ARS2+/− and _ARS2_−/− E3.5 embryos before and after 24 and 48 h of culture. Before culture (t = 0), the nuclear localization of ARS2 was detected with anti-ARS2 polyclonal antibody and Alexa Fluor 568 goat anti-rabbit IgG (red) in the wild type and was present in smaller amounts or even absent in null embryos. Nuclear DNA was stained using DRAQ5 (blue); the merge of red and blue and the differential interference microscopy (DIC) are indicated. Embryos were observed by confocal microscopy, and a representative single optical section is shown. At time zero, the arrow indicates the nuclear localization of ARS2 in a cell undergoing anaphase. After 24 h of culture (t = 24), both ARS2+/− and _ARS2_−/− embryos had formed expanded blastocysts and _ARS2_−/− embryos had completely lost their nuclear ARS2 localization. After 48 h of culture (t = 48), the null blastocyst had collapsed and several abnormal nuclei could be observed. The scale bars correspond to 50 μm.
FIG. 7.
Absence of blastocyst outgrowths in _ARS2_−/− embryos. Representative examples of cultured E3.5 blastocysts from ARS2+/− intercrosses are shown. The embryos were photographed every 24 h using phase-contrast microscopy. All embryos were genotyped after being cultured. At E3.5, no obvious differences between ARS2+/+ ARS2+/− and _ARS2_−/− embryos were observed. By E3.5 plus 24 h (E3.5 + 1), some _ARS2_−/− embryos had collapsed and were generally not attached to the culture dish. ARS2+/+ and ARS2+/− embryos had both expanded and attached by this time. By 48 h (E3.5 + 2), ARS2+/+ and ARS2+/− embryos hatched from their zona pellucida (ZP), whereas _ARS2_−/− embryos invariably failed to hatch and were normally found floating in the medium or loosely attached to the gelatin-coated plates. The removal of the ZP in null blastocysts did not affect the phenotype (column 4). After 120 h (E3.5 + 5) of culture, ARS2+/+ and ARS2+/− embryos were composed of outgrowths of ICM and were surrounded by a single layer of trophectoderm giant cells (TGC), whereas null embryos remained a small cluster of abnormal cells showing severe membrane blebbing (arrows).
FIG. 8.
Localization of the lineage-specific markers Cdx2 and Oct4 in ARS2+/+ and _ARS2_−/− embryos. The trophectoderm marker Cdx2 was labeled with anti-CDX2 and Alexa Fluor 488 goat anti-mouse IgG (green). The ICM marker Oct4 was labeled with anti-OCT4 and Alexa Fluor 568 goat anti-rabbit IgG (red). Cells without either label showing condensed nuclei are indicated by arrows. The nuclei were labeled with DRAQ5 (blue). The embryos were observed by confocal microscopy, and a representative single optical section is shown.
FIG. 9.
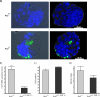
Cultured _ARS2_-null embryos show excessive apoptosis. E3.5 blastocysts were cultured for 24 h, apoptotic cells were identified using a fluorescent TUNEL assay (green), and morphology was assessed with DRAQ5 nuclear stain (blue). The embryos were genotyped by PCR (ARS2+/+ ARS2+/−, n = 12; _ARS2_−/−, n = 7). (A) A representative _ARS2_−/− embryo showing excessive TUNEL labeling predominantly in the trophectoderm and an ARS2+/− littermate. (B) There was a significant increase in TUNEL-positive nuclei in _ARS2_−/− over ARS2+/+ ARS2+/− embryos (_t_[28] = 7.4; P < 0.0001). (C) Cell numbers in E3.5 embryos cultured for 24 h showed a statistically significant decrease in _ARS2_−/− cell numbers over ARS2+/+ ARS2+/− E3.5 blastocysts cultured for 24 h (_t_[38] = 2.3; P = 0.0283). (D) No statistically significant difference in the mitotic index was observed between ARS2+/+ ARS2+/− (n = 12) and _ARS2_−/− embryos (_t_[10] = 1.45; P = 0.177).
Similar articles
- Taube nuss is a novel gene essential for the survival of pluripotent cells of early mouse embryos.
Voss AK, Thomas T, Petrou P, Anastassiadis K, Schöler H, Gruss P. Voss AK, et al. Development. 2000 Dec;127(24):5449-61. doi: 10.1242/dev.127.24.5449. Development. 2000. PMID: 11076765 - Mutagenesis of ARS2 Domains To Assess Possible Roles in Cell Cycle Progression and MicroRNA and Replication-Dependent Histone mRNA Biogenesis.
O'Sullivan C, Christie J, Pienaar M, Gambling J, Nickerson PE, Alford SC, Chow RL, Howard PL. O'Sullivan C, et al. Mol Cell Biol. 2015 Nov;35(21):3753-67. doi: 10.1128/MCB.00272-15. Epub 2015 Aug 24. Mol Cell Biol. 2015. PMID: 26303529 Free PMC article. - Gene structure and embryonic expression of mouse COP9 signalosome subunit 8 (Csn8).
Lykke-Andersen K, Wei N. Lykke-Andersen K, et al. Gene. 2003 Dec 4;321:65-72. doi: 10.1016/s0378-1119(03)00836-9. Gene. 2003. PMID: 14636993 - ARS2/SRRT: at the nexus of RNA polymerase II transcription, transcript maturation and quality control.
Lykke-Andersen S, Rouvière JO, Jensen TH. Lykke-Andersen S, et al. Biochem Soc Trans. 2021 Jun 30;49(3):1325-1336. doi: 10.1042/BST20201008. Biochem Soc Trans. 2021. PMID: 34060620 Review. - Arsenic resistance protein 2 and microRNA biogenesis: Biological implications in cancer development.
Yuan L, Jiang X, Gong Q, Gao N. Yuan L, et al. Pharmacol Ther. 2023 Apr;244:108386. doi: 10.1016/j.pharmthera.2023.108386. Epub 2023 Mar 16. Pharmacol Ther. 2023. PMID: 36933704 Review.
Cited by
- Cytoplasmic switch of ARS2 isoforms promotes nonsense-mediated mRNA decay and arsenic sensitivity.
Mesa-Perez M, Hamilton PT, Miranda A, Brodie N, O'Sullivan C, Christie J, Ryan BC, Chow RL, Goodlett D, Nelson CJ, Howard PL. Mesa-Perez M, et al. Nucleic Acids Res. 2022 Feb 22;50(3):1620-1638. doi: 10.1093/nar/gkac033. Nucleic Acids Res. 2022. PMID: 35104878 Free PMC article. - miRNA biogenesis: biological impact in the development of cancer.
Romero-Cordoba SL, Salido-Guadarrama I, Rodriguez-Dorantes M, Hidalgo-Miranda A. Romero-Cordoba SL, et al. Cancer Biol Ther. 2014;15(11):1444-55. doi: 10.4161/15384047.2014.955442. Cancer Biol Ther. 2014. PMID: 25482951 Free PMC article. Review. - Identification of Three Key Genes Associated with Hepatocellular Carcinoma Progression Based on Co-expression Analysis.
Lin J, Zhang F. Lin J, et al. Cell Biochem Biophys. 2022 Jun;80(2):301-309. doi: 10.1007/s12013-021-01028-2. Epub 2021 Aug 18. Cell Biochem Biophys. 2022. PMID: 34406599 - Microprocessor of microRNAs: regulation and potential for therapeutic intervention.
Beezhold KJ, Castranova V, Chen F. Beezhold KJ, et al. Mol Cancer. 2010 Jun 1;9:134. doi: 10.1186/1476-4598-9-134. Mol Cancer. 2010. PMID: 20515486 Free PMC article. Review. - Arabidopsis RNA processing factor SERRATE regulates the transcription of intronless genes.
Speth C, Szabo EX, Martinho C, Collani S, Zur Oven-Krockhaus S, Richter S, Droste-Borel I, Macek B, Stierhof YD, Schmid M, Liu C, Laubinger S. Speth C, et al. Elife. 2018 Aug 28;7:e37078. doi: 10.7554/eLife.37078. Elife. 2018. PMID: 30152752 Free PMC article.
References
- Clarke, J. H., D. Tack, K. Findlay, M. Van Montagu, and M. Van Lijsebettens. 1999. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 20493-501. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases