Canine population structure: assessment and impact of intra-breed stratification on SNP-based association studies - PubMed (original) (raw)
Canine population structure: assessment and impact of intra-breed stratification on SNP-based association studies
Pascale Quignon et al. PLoS One. 2007.
Abstract
Background: In canine genetics, the impact of population structure on whole genome association studies is typically addressed by sampling approximately equal numbers of cases and controls from dogs of a single breed, usually from the same country or geographic area. However one way to increase the power of genetic studies is to sample individuals of the same breed but from different geographic areas, with the expectation that independent meiotic events will have shortened the presumed ancestral haplotype around the mutation differently. Little is known, however, about genetic variation among dogs of the same breed collected from different geographic regions.
Methodology/principal findings: In this report, we address the magnitude and impact of genetic diversity among common breeds sampled in the U.S. and Europe. The breeds selected, including the Rottweiler, Bernese mountain dog, flat-coated retriever, and golden retriever, share susceptibility to a class of soft tissue cancers typified by malignant histiocytosis in the Bernese mountain dog. We genotyped 722 SNPs at four unlinked loci (between 95 and 271 per locus) on canine chromosome 1 (CFA1). We showed that each population is characterized by distinct genetic diversity that can be correlated with breed history. When the breed studied has a reduced intra-breed diversity, the combination of dogs from international locations does not increase the rate of false positives and potentially increases the power of association studies. However, over-sampling cases from one geographic location is more likely to lead to false positive results in breeds with significant genetic diversity.
Conclusions: These data provide new guidelines for association studies using purebred dogs that take into account population structure.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
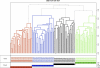
Figure 1. Hierarchical clustering between the four breeds.
Clustering of the 119 dogs (k = 4) from the four different breeds. In the dendogram each vertical line represents a single dog and the color reflects the four clusters obtained by PLINK analysis. A grey dashed line indicates an outlier dog. Below the dendogram, dogs are named by their breed origin, and names are colored upon their cluster assignment. At the bottom of the figure, colored squares are drawn on four lines, each line representing the different breeds. The scale on the right axis represents the genetics distances calculated by PLINK software. Each breed separates from the others as the four colors correspond exactly to the four breeds, red for BMD, blue for FCR, black for GR and green for ROT.
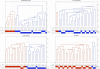
Figure 2. Hierarchical clustering within breeds.
The figure shows the clustering analysis (k = 2) within each breed for BMD, FCR, ROT and GR (n = 30 for each breed). For each breed, a dendogram is drawn in which each vertical line represents a single dog. The two clusters are represented in blue and red, and grey dashed lines indicate outlier dogs. Below the dendogram, dogs are named by geographical origin (US or EU), and names are colored upon their cluster assignment. The scale on the right axis represents the genetics distances calculated by PLINK software.
Similar articles
- Extensive and breed-specific linkage disequilibrium in Canis familiaris.
Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, Kruglyak L, Ostrander EA. Sutter NB, et al. Genome Res. 2004 Dec;14(12):2388-96. doi: 10.1101/gr.3147604. Epub 2004 Nov 15. Genome Res. 2004. PMID: 15545498 Free PMC article. - Assessment of the functionality of genome-wide canine SNP arrays and implications for canine disease association studies.
Ke X, Kennedy LJ, Short AD, Seppälä EH, Barnes A, Clements DN, Wood SH, Carter SD, Happ GM, Lohi H, Ollier WE. Ke X, et al. Anim Genet. 2011 Apr;42(2):181-90. doi: 10.1111/j.1365-2052.2010.02132.x. Epub 2010 Nov 11. Anim Genet. 2011. PMID: 21070295 - Extent of linkage disequilibrium in large-breed dogs: chromosomal and breed variation.
Stern JA, White SN, Meurs KM. Stern JA, et al. Mamm Genome. 2013 Oct;24(9-10):409-15. doi: 10.1007/s00335-013-9474-y. Epub 2013 Sep 6. Mamm Genome. 2013. PMID: 24062056 - Systematically identifying genetic signatures including novel SNP-clusters, nonsense variants, frame-shift INDELs, and long STR expansions that potentially link to unknown phenotypes existing in dog breeds.
Li Z, Wang Z, Chen Z, Voegeli H, Lichtman JH, Smith P, Liu J, DeWan AT, Hoh J. Li Z, et al. BMC Genomics. 2023 Jun 5;24(1):302. doi: 10.1186/s12864-023-09390-6. BMC Genomics. 2023. PMID: 37277710 Free PMC article. - Forensic DNA phenotyping: Canis familiaris breed classification and skeletal phenotype prediction using functionally significant skeletal SNPs and indels.
Raymond PW, Velie BD, Wade CM. Raymond PW, et al. Anim Genet. 2022 Jun;53(3):247-263. doi: 10.1111/age.13165. Epub 2021 Dec 28. Anim Genet. 2022. PMID: 34963196 Review.
Cited by
- Assessing breed integrity of Göttingen Minipigs.
Reimer C, Ha NT, Sharifi AR, Geibel J, Mikkelsen LF, Schlather M, Weigend S, Simianer H. Reimer C, et al. BMC Genomics. 2020 Apr 16;21(1):308. doi: 10.1186/s12864-020-6590-4. BMC Genomics. 2020. PMID: 32299342 Free PMC article. - Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development.
Parker HG, Dreger DL, Rimbault M, Davis BW, Mullen AB, Carpintero-Ramirez G, Ostrander EA. Parker HG, et al. Cell Rep. 2017 Apr 25;19(4):697-708. doi: 10.1016/j.celrep.2017.03.079. Cell Rep. 2017. PMID: 28445722 Free PMC article. - Identification of genetic variation and haplotype structure of the canine ABCA4 gene for retinal disease association studies.
Zangerl B, Lindauer SJ, Acland GM, Aguirre GD. Zangerl B, et al. Mol Genet Genomics. 2010 Oct;284(4):243-50. doi: 10.1007/s00438-010-0560-5. Epub 2010 Jul 27. Mol Genet Genomics. 2010. PMID: 20661590 Free PMC article. - Real-time PCR genotyping assay for GM2 gangliosidosis variant 0 in toy poodles and the mutant allele frequency in Japan.
Rahman MM, Yabuki A, Kohyama M, Mitani S, Mizukami K, Uddin MM, Chang HS, Kushida K, Kishimoto M, Yamabe R, Yamato O. Rahman MM, et al. J Vet Med Sci. 2014 Mar 1;76(2):295-9. doi: 10.1292/jvms.13-0443. Epub 2013 Oct 25. J Vet Med Sci. 2014. PMID: 24161966 Free PMC article.
References
- Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. - PubMed
- Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005;15:1706–1716. - PubMed
- Galibert F, André C. [The dog and its genome]. Med Sci (Paris) 2006;22:806–808. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources