The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi - PubMed (original) (raw)
The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi
I Hernández-Lucas et al. J Bacteriol. 2008 Mar.
Abstract
LeuO is a LysR-type transcriptional regulator that has been implicated in the bacterial stringent response and in the virulence of Salmonella. A genomic analysis with Salmonella enterica serovar Typhi revealed that LeuO is a positive regulator of OmpS1, OmpS2, AssT, and STY3070. In contrast, LeuO down-regulated the expression of OmpX, Tpx, and STY1978. Transcriptional fusions supported the positive and negative LeuO regulation. Expression of ompS1, assT, and STY3070 was induced in an hns mutant, consistent with the notion that H-NS represses these genes; transcriptional activity was lower for tpx and STY1978 in an hns background, suggesting that this global regulatory protein has a positive effect. In contrast, ompS2 and ompX expression appeared to be H-NS independent. LeuO specifically bound to the 5' intergenic regions of ompS2, assT, STY3070, ompX, and tpx, while it was not observed to bind to the promoter region of STY1978, suggesting that LeuO regulates in direct and indirect ways. In this work, a novel set of genes belonging to the LeuO regulon are described; interestingly, these genes are involved in a variety of biological processes, suggesting that LeuO is a global regulator in Salmonella.
Figures
FIG. 1.
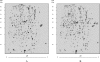
Proteomic profiles of Salmonella serovar Typhi total protein extracts. (A) Salmonella serovar Typhi Δ_leuO_ containing pFM_Trc_12 (IMSS-32). (B) Salmonella serovar Typhi containing pFM_TrcleuO_-50 (IMSS-III). Cells were grown in MA broth to an OD595 of 0.6. Cultures were supplemented with 50 μM IPTG. The labeled spots were excised and identified using MALDI-TOF MS. The identities of circled proteins are shown in Table 1. More than three independent experiments were performed, and representative 2-DGE gels are shown.
FIG. 2.
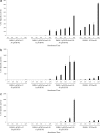
Transcriptional profiles of the LeuO-dependent genes fused to the cat reporter gene. Bacterial strains IMSS-II (Salmonella serovar Typhi IMSS-1 harboring plasmid pFM_Trc_12) and IMSS-III (Salmonella serovar Typhi IMSS-1 harboring plasmid pFM_TrcleuO_-50) were independently transformed with fusions of each of the LeuO-regulated genes (pKK232-9 [STY3070], pKK232-9 [_assT_], pKK232-9 [_ompS1_], pKK232-9 [_ompS2_], pKK232-9 [_tpx_], pKK232-9 [_ompX_], and pKK232-9 [STY1978]) to generate individual strains containing the LeuO regulator, as well as the promoter region of the LeuO-dependent gene. To evaluate the effect of H-NS in the regulation of each of the LeuO-regulated genes, individual fusions were transformed into Salmonella serovar Typhi hns mutant STY_hns_99. The CAT specific activity of each strain was determined using samples collected at OD595 of 0.4, 0.6, 0.8, and 1 and at 12 h. For strains IMSS-II and IMSS-III 50 μM IPTG was used as an inducer. The bars indicate the means of three independent experiments performed in duplicate.
FIG. 2.
Transcriptional profiles of the LeuO-dependent genes fused to the cat reporter gene. Bacterial strains IMSS-II (Salmonella serovar Typhi IMSS-1 harboring plasmid pFM_Trc_12) and IMSS-III (Salmonella serovar Typhi IMSS-1 harboring plasmid pFM_TrcleuO_-50) were independently transformed with fusions of each of the LeuO-regulated genes (pKK232-9 [STY3070], pKK232-9 [_assT_], pKK232-9 [_ompS1_], pKK232-9 [_ompS2_], pKK232-9 [_tpx_], pKK232-9 [_ompX_], and pKK232-9 [STY1978]) to generate individual strains containing the LeuO regulator, as well as the promoter region of the LeuO-dependent gene. To evaluate the effect of H-NS in the regulation of each of the LeuO-regulated genes, individual fusions were transformed into Salmonella serovar Typhi hns mutant STY_hns_99. The CAT specific activity of each strain was determined using samples collected at OD595 of 0.4, 0.6, 0.8, and 1 and at 12 h. For strains IMSS-II and IMSS-III 50 μM IPTG was used as an inducer. The bars indicate the means of three independent experiments performed in duplicate.
FIG. 2.
Transcriptional profiles of the LeuO-dependent genes fused to the cat reporter gene. Bacterial strains IMSS-II (Salmonella serovar Typhi IMSS-1 harboring plasmid pFM_Trc_12) and IMSS-III (Salmonella serovar Typhi IMSS-1 harboring plasmid pFM_TrcleuO_-50) were independently transformed with fusions of each of the LeuO-regulated genes (pKK232-9 [STY3070], pKK232-9 [_assT_], pKK232-9 [_ompS1_], pKK232-9 [_ompS2_], pKK232-9 [_tpx_], pKK232-9 [_ompX_], and pKK232-9 [STY1978]) to generate individual strains containing the LeuO regulator, as well as the promoter region of the LeuO-dependent gene. To evaluate the effect of H-NS in the regulation of each of the LeuO-regulated genes, individual fusions were transformed into Salmonella serovar Typhi hns mutant STY_hns_99. The CAT specific activity of each strain was determined using samples collected at OD595 of 0.4, 0.6, 0.8, and 1 and at 12 h. For strains IMSS-II and IMSS-III 50 μM IPTG was used as an inducer. The bars indicate the means of three independent experiments performed in duplicate.
FIG. 3.
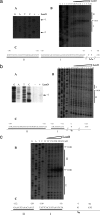
Transcriptional start sites and LeuO protected regions of genes belonging to the LeuO regulon. Primer extension and DNase I protection assays for the STY3070, assT ompS2, ompX, and tpx genes were performed. For STY1978, only the transcriptional initiation results are shown. (A) Transcription initiation in the presence (+) or absence (−) of LeuO. (B) LeuO sites protected from DNase I. The LeuO concentrations used are indicated at the top. D− and D+ indicate without and with DNase I, respectively. DHR, DNase I hypersensitive regions. (C) Schematic diagram of the regulatory region of the LeuO-dependent genes. The nucleotides corresponding to the transcription initiation site ( ) the LeuO regions protected from DNase I (I and II), and the translation initiation sites (ATG) are indicated.
) the LeuO regions protected from DNase I (I and II), and the translation initiation sites (ATG) are indicated.
FIG. 3.
Transcriptional start sites and LeuO protected regions of genes belonging to the LeuO regulon. Primer extension and DNase I protection assays for the STY3070, assT ompS2, ompX, and tpx genes were performed. For STY1978, only the transcriptional initiation results are shown. (A) Transcription initiation in the presence (+) or absence (−) of LeuO. (B) LeuO sites protected from DNase I. The LeuO concentrations used are indicated at the top. D− and D+ indicate without and with DNase I, respectively. DHR, DNase I hypersensitive regions. (C) Schematic diagram of the regulatory region of the LeuO-dependent genes. The nucleotides corresponding to the transcription initiation site ( ) the LeuO regions protected from DNase I (I and II), and the translation initiation sites (ATG) are indicated.
) the LeuO regions protected from DNase I (I and II), and the translation initiation sites (ATG) are indicated.
Similar articles
- OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene.
Fernández-Mora M, Puente JL, Calva E. Fernández-Mora M, et al. J Bacteriol. 2004 May;186(10):2909-20. doi: 10.1128/JB.186.10.2909-2920.2004. J Bacteriol. 2004. PMID: 15126450 Free PMC article. - LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1.
De la Cruz MA, Fernández-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vázquez A, Calva E. De la Cruz MA, et al. Mol Microbiol. 2007 Nov;66(3):727-43. doi: 10.1111/j.1365-2958.2007.05958.x. Epub 2007 Oct 1. Mol Microbiol. 2007. PMID: 17908208 - Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella typhi: a novel regulatory mechanism that involves OmpR.
Oropeza R, Sampieri CL, Puente JL, Calva E. Oropeza R, et al. Mol Microbiol. 1999 Apr;32(2):243-52. doi: 10.1046/j.1365-2958.1999.01329.x. Mol Microbiol. 1999. PMID: 10231482 - The Subtleties and Contrasts of the LeuO Regulator in Salmonella Typhi: Implications in the Immune Response.
Guadarrama C, Villaseñor T, Calva E. Guadarrama C, et al. Front Immunol. 2014 Dec 12;5:581. doi: 10.3389/fimmu.2014.00581. eCollection 2014. Front Immunol. 2014. PMID: 25566242 Free PMC article. Review. - The LeuO regulator and quiescence: About transcriptional roadblocks, multiple promoters, and crispr-cas.
Sánchez-Popoca D, Serrano-Fujarte I, Fernández-Mora M, Calva E. Sánchez-Popoca D, et al. Mol Microbiol. 2022 Nov;118(5):503-509. doi: 10.1111/mmi.14990. Epub 2022 Oct 20. Mol Microbiol. 2022. PMID: 36203248 Review.
Cited by
- The Salmonella enterica serovar Typhi LeuO global regulator forms tetramers: residues involved in oligomerization, DNA binding, and transcriptional regulation.
Guadarrama C, Medrano-López A, Oropeza R, Hernández-Lucas I, Calva E. Guadarrama C, et al. J Bacteriol. 2014 Jun;196(12):2143-54. doi: 10.1128/JB.01484-14. Epub 2014 Mar 21. J Bacteriol. 2014. PMID: 24659766 Free PMC article. - Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems.
Richter C, Chang JT, Fineran PC. Richter C, et al. Viruses. 2012 Oct 19;4(10):2291-311. doi: 10.3390/v4102291. Viruses. 2012. PMID: 23202464 Free PMC article. Review. - I can see CRISPR now, even when phage are gone: a view on alternative CRISPR-Cas functions from the prokaryotic envelope.
Ratner HK, Sampson TR, Weiss DS. Ratner HK, et al. Curr Opin Infect Dis. 2015 Jun;28(3):267-74. doi: 10.1097/QCO.0000000000000154. Curr Opin Infect Dis. 2015. PMID: 25887612 Free PMC article. Review. - The Salmonella enterica Serovar Typhi ltrR Gene Encodes Two Proteins Whose Transcriptional Expression Is Upregulated by Alkaline pH and Repressed at Their Promoters and Coding Regions by H-NS and Lrp.
Rebollar-Flores JE, Medina-Aparicio L, Osio-Becerro VE, Villarreal JM, Mayo S, Mendoza BD, Rodríguez-Gutierrez S, Olvera L, Dávila S, Encarnación S, Martínez-Batallar AG, Calva E, Hernández-Lucas I. Rebollar-Flores JE, et al. J Bacteriol. 2020 Jun 9;202(13):e00783-19. doi: 10.1128/JB.00783-19. Print 2020 Jun 9. J Bacteriol. 2020. PMID: 32284321 Free PMC article. - NhaR, LeuO, and H-NS Are Part of an Expanded Regulatory Network for Ectoine Biosynthesis Expression.
Boas Lichty KE, Gregory GJ, Boyd EF. Boas Lichty KE, et al. Appl Environ Microbiol. 2023 Jun 28;89(6):e0047923. doi: 10.1128/aem.00479-23. Epub 2023 Jun 6. Appl Environ Microbiol. 2023. PMID: 37278653 Free PMC article.
References
- Chen, C. C., M. Y. Chou, C. H. Huang, A. Majumder, and H. Y. Wu. 2005. A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J. Biol. Chem. 2805101-5112. - PubMed
- Chen, C. C., and H. Y. Wu. 2005. LeuO protein delimits the transcriptionally active and repressive domains on the bacterial chromosome. J. Biol. Chem. 28015111-15121. - PubMed
- Chen, C. C., M. Ghole, A. Majumder, Z. Wang, S. Chandana, and H. Y. Wu. 2003. LeuO-mediated transcriptional derepression. J. Biol. Chem. 27838094-38103. - PubMed
- De la Cruz, M. A., M. Fernández-Mora, C. Guadarrama, M. A Flores-Valdez, V. H. Bustamante, A. Vázquez, and E. Calva. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol. Microbiol. 66727-743. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous