Experimental validation of miRNA targets - PubMed (original) (raw)
Review
Experimental validation of miRNA targets
Donald E Kuhn et al. Methods. 2008 Jan.
Abstract
MicroRNAs are natural, single-stranded, small RNA molecules that regulate gene expression by binding to target mRNAs and suppress its translation or initiate its degradation. In contrast to the identification and validation of many miRNA genes is the lack of experimental evidence identifying their corresponding mRNA targets. The most fundamental challenge in miRNA biology is to define the rules of miRNA target recognition. This is critical since the biological role of individual miRNAs will be dictated by the mRNAs that they regulate. Therefore, only as target mRNAs are validated will it be possible to establish commonalities that will enable more precise predictions of miRNA/mRNA interactions. Currently there is no clear agreement as to what experimental procedures should be followed to demonstrate that a given mRNA is a target of a specific miRNA. Therefore, this review outlines several methods by which to validate miRNA targets. Additionally, we propose that multiple criteria should be met before miRNA target validation should be considered "confirmed."
Figures
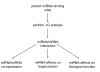
Figure 1
Proposed flow diagram for microRNA validation.
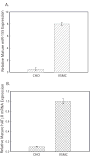
Figure 2
miR-155 can specifically inhibit luciferase reporter activity. CHO cells were cotransfected with pMIR/883/5′→3′, pMIR/883/3′→5′, pMIR/883/5′→3′/ΔmiR-155, pRL-CMV and 50 nM of a given miRNA mimic. Forty-eight hours after transfection luciferase activities were measured. F-luc activity was normalized to r-luc expression and the mean activities ± S.E. from five independent experiments are shown. (*p<0.01 versus CHO cells transfected with miR-control mimic)
Figure 3
Mature miR-155 and the hAT1R are co-expressed in VSMC. The TaqMan® miR-155 and TaqMan® AT1R gene expression assay kits were utilized to quantify the expression of miR-155 and hAT1R levels in VSM and CHO cells. The expression of mature miR-155 or hAT1R relative to 18S rRNA was determined using SYBR green real-time quantitative PCR assay as described [21].
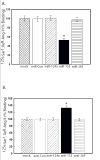
Figure 4
miR-155 is expressed in endothelial and VSM cells. A. Representative example of the distribution of mature miR-155 after in situ hybridization analysis with an LNA miR-155-specific antisense probe. In the arteriole, the endothelial (small arrow) and VSM (large arrow) cells abundantly express miR-155. B. No signal was evident in the arteriole when the serial section was probed with a scrambled probe.
Figure 5
miR-155 regulates the endogenous hAT1R expression in VSMCs. VSMCs were either mock transfected or transfected with the miRNA mimics (A) or antisense miRNAs (B) as indicated. 48 hrs after transfection, the cells were utilized for AT1R radioreceptor binding assays as described [21]. The data have been normalized for protein and transfection differences and represent specific binding. The values are shown as percent of maximal specific binding of mock transfected hPFBs and represent the mean ± S.E. from four independent experiments (*p<0.001 vs. mock transfected cells).
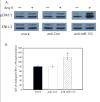
Figure 6
Anti-miR-155 enhances Ang II-induced signaling in VSMCs. VSMCs were transfected with the anti-miRNA oligonucleotides as indicated. A. Ang II-induced phosphoERK1/2 experiments were performed utilizing serum-starved, transiently-transfected cells as described [21,22]. A representative immunoblot is shown. Results are representative of four independent experiments. B. The quantitation of Ang II-(1 μM for 5 min) induced ERK1/2 phosphorylation was determined by densitometry. Values are expressed as a percent of the maximal phosphorylation of ERK1/2 in response to Ang II in mock transfected cells and represents the mean ± S.E. from four independent transfection experiments (*p<0.01).
Similar articles
- Identification and validation of plant miRNA from NGS data-an experimental approach.
Devi K, Dey KK, Singh S, Mishra SK, Modi MK, Sen P. Devi K, et al. Brief Funct Genomics. 2019 Feb 14;18(1):13-22. doi: 10.1093/bfgp/ely034. Brief Funct Genomics. 2019. PMID: 30335137 Review. - Experimental procedures to identify and validate specific mRNA targets of miRNAs.
Elton TS, Yalowich JC. Elton TS, et al. EXCLI J. 2015 Jul 2;14:758-90. doi: 10.17179/excli2015-319. eCollection 2015. EXCLI J. 2015. PMID: 27047316 Free PMC article. Review. - A study of miRNAs targets prediction and experimental validation.
Huang Y, Zou Q, Song H, Song F, Wang L, Zhang G, Shen X. Huang Y, et al. Protein Cell. 2010 Nov;1(11):979-86. doi: 10.1007/s13238-010-0129-4. Epub 2010 Dec 10. Protein Cell. 2010. PMID: 21153515 Free PMC article. Review. - Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets.
Wang XJ, Reyes JL, Chua NH, Gaasterland T. Wang XJ, et al. Genome Biol. 2004;5(9):R65. doi: 10.1186/gb-2004-5-9-r65. Epub 2004 Aug 31. Genome Biol. 2004. PMID: 15345049 Free PMC article. - Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data.
Wang YP, Li KB. Wang YP, et al. BMC Genomics. 2009 May 12;10:218. doi: 10.1186/1471-2164-10-218. BMC Genomics. 2009. PMID: 19435500 Free PMC article.
Cited by
- Improving miRNA-mRNA interaction predictions.
Tabas-Madrid D, Muniategui A, Sánchez-Caballero I, Martínez-Herrera DJ, Sorzano CO, Rubio A, Pascual-Montano A. Tabas-Madrid D, et al. BMC Genomics. 2014;15 Suppl 10(Suppl 10):S2. doi: 10.1186/1471-2164-15-S10-S2. Epub 2014 Dec 12. BMC Genomics. 2014. PMID: 25559987 Free PMC article. - Impact of host genes and strand selection on miRNA and miRNA* expression.
Biasiolo M, Sales G, Lionetti M, Agnelli L, Todoerti K, Bisognin A, Coppe A, Romualdi C, Neri A, Bortoluzzi S. Biasiolo M, et al. PLoS One. 2011;6(8):e23854. doi: 10.1371/journal.pone.0023854. Epub 2011 Aug 31. PLoS One. 2011. PMID: 21909367 Free PMC article. - MicroRNAs and prostate cancer.
Shi XB, Tepper CG, White RW. Shi XB, et al. J Cell Mol Med. 2008 Sep-Oct;12(5A):1456-65. doi: 10.1111/j.1582-4934.2008.00420.x. Epub 2008 Jul 8. J Cell Mol Med. 2008. PMID: 18624768 Free PMC article. Review. - MicroRNA target prediction by expression analysis of host genes.
Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. Gennarino VA, et al. Genome Res. 2009 Mar;19(3):481-90. doi: 10.1101/gr.084129.108. Epub 2008 Dec 16. Genome Res. 2009. PMID: 19088304 Free PMC article. - MET as a target for treatment of chest tumors.
Cipriani NA, Abidoye OO, Vokes E, Salgia R. Cipriani NA, et al. Lung Cancer. 2009 Feb;63(2):169-79. doi: 10.1016/j.lungcan.2008.06.011. Epub 2008 Jul 30. Lung Cancer. 2009. PMID: 18672314 Free PMC article. Review.
References
- Bartel DP. Cell. 2004;116:281–297. - PubMed
- Bushati N, Cohen SM. Ann Review of Cell Develop Biol. 2007;23:175–205. - PubMed
- Wang Y, Stricker HM, Gou O, Liu L. Frontiers in Bioscience. 2007;12:2316–2329. - PubMed
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzila A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Nat Genet. 2005;37:766–770. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous