Architecture of a coat for the nuclear pore membrane - PubMed (original) (raw)
Architecture of a coat for the nuclear pore membrane
Kuo-Chiang Hsia et al. Cell. 2007.
Abstract
The symmetric core of the nuclear pore complex can be considered schematically as a series of concentric cylinders. A peripheral cylinder coating the pore membrane contains the previously characterized, elongated heptamer that harbors Sec13-Nup145C in its middle section. Strikingly, Sec13-Nup145C crystallizes as a hetero-octamer in two space groups. Oligomerization of Sec13-Nup145C was confirmed biochemically. Importantly, the numerous interacting surfaces in the hetero-octamer are evolutionarily highly conserved, further underlining the physiological relevance of the oligomerization. The hetero-octamer forms a slightly curved, yet rigid rod of sufficient length to span the entire height of the proposed membrane-adjacent cylinder. In concordance with the dimensions and symmetry of the nuclear pore complex core, we suggest that the cylinder is constructed of four antiparallel rings, each ring being composed of eight heptamers arranged in a head-to-tail fashion. Our model proposes that the hetero-octamer would vertically traverse and connect the four stacked rings.
Figures
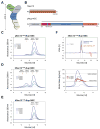
Figure 1. Organization and Dynamic Behavior of the Sec13-Nup145C Complex
(A) Schematic representation of the heptameric complex and the approximate localization of its seven nups (Lutzmann et al., 2002). (B) Domain structures of human Sec13 and yeast Nup145C. For Sec13, the WD40 repeats (orange), the C-terminal tail (T) (red), and the numbering relative to human Sec13 are indicated. For Nup145C, the unstructured N-terminal region (gray), the domain invasion motif (DIM) (purple), the αB-αC connector (C) (red), the α-helical domain (blue), and the C-terminal α-helical region (orange) are indicated. The numbering is relative to the yeast Nup145C. The bars above the domain structures of both proteins mark the crystallized fragments and are referred to as the Sec13-Nup145C complex. Gel filtration profiles of (C) the ySec13-Nup145C complex, (D) the hSec131–322-Nup145C complex, and (E) the C-terminally truncated hSec131–316-Nup145C complex. All proteins were injected at the indicated concentrations. The predicted elution positions for the various assembly states of the Sec13-Nup145C complexes are shown and have been determined using molecular weight standards. (F) Multi-angle light scattering analysis of the hSec131–316-Nup145C complex. The black bars on each side of the peak delimit the data points used for further analysis (upper panel). Molecular weights were determined by light scattering at the three indicated protein concentrations (lower panel). The complex forms a concentration-dependent, polydisperse dynamic equilibrium between hetero-dimeric and hetero-tetrameric assemblies in solution.
Figure 2. Overview of the Structure of the Sec13-Nup145C Hetero-Octamer
(A) Ribbon representation of the Sec13-Nup145C hetero-octamer, showing Sec13 in yellow and orange and Nup145C in green and blue. A 90° rotated view is shown on the right. The three pseudo-two-fold axes (black ovals) that run through the hetero-octamer and the overall dimensions are indicated. The Sec13-Nup145C hetero-octamer forms a slightly bent rod. (B) Schematic representation of the Sec13-Nup145C hetero-octamer. Magenta lines indicate interaction surfaces.
Figure 3. The Interaction of the Nup145C α-Helical Domain with the Sec13 β-Propeller
(A) The ribbon representation of the Nup145C structure is shown in rainbow colors along the polypeptide chain from the N- to the C-terminus. The N-terminal domain invasion motif (DIM), the C-terminal α-helical domain, and their secondary structure elements are indicated. (B) The structure of the Sec13-Nup145C hetero-dimer. The Nup145CDIM (magenta), the Nup145C α-helical domain (blue), the Nup145C αB-αC connector segment (red), and the Sec13 β-propeller (yellow) are indicated; a 90° rotated view is shown on the right. (C) Schematic representation of the Sec13-Nup145C interaction. (D) The β-propeller domain of Sec13 in complex with the Nup145CDIM. Sec13 is shown in yellow, and the six blades are indicated. The Nup145CDIM forms a three-stranded seventh blade, complementing the Sec13 β-propeller domain. (E) Schematic representation of the Sec13 β-propeller and its interaction with the Nup145CDIM.
Figure 4. Surface Properties of the Sec13-Nup145C Hetero-Dimer
(A) Surface rendition of the Sec13-Nup145C complex. The surface is colored according to the proteins (Sec13, yellow; Nup145C, blue) and their participation in various interactions (Sec13 from the adjacent complex, orange; Nup145C from the adjacent complex, purple; Nup145C homo-dimerization, green). (B) Nup145C is colored according to sequence conservation, from 40 % similarity (white) to 100 % identity (green). (C) Surface rendition of Nup145C, colored according to the electrostatic potential, from red (−10 kBT/e) to blue (+10 kBT/e).
Figure 5. The Assembly of the Sec13-Nup145C Hetero-Octamer
(A) Ribbon representation of the dimerization of the Nup145C α-helical domain. For clarity, only the two interacting Nup145C protomers in the hetero-octamer are colored (green and blue). The centrally located α-helices that facilitate the homo-dimerization of Nup145C are indicated. A 180° rotated view is shown on the right. (B) The dimerization interface located in the center of the Sec13-Nup145C hetero-octamer. The two Sec13 β-propeller domains (orange and yellow) and two Nup145C molecules (blue) are colored. The two Sec13 C-termini (the last ordered residue is Val304) are indicated and are only ~10 Å apart. A 90° rotated view is shown below. The locations of the pseudo-two-fold axes of symmetry that run through both interfaces are indicated (black ovals).
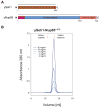
Figure 6. Biochemical Characterization of the Seh1-Nup85 Complex
(A) Domain structures of yeast Seh1 and yeast Nup85. For Seh1, the six WD40 repeats (orange), the C-terminal tail (T) (red), and the numbering are indicated. For Nup85, the predicted domain invasion motif (DIM) (magenta), the 〈-helical domain (blue), and the C-terminal 〈-helical region (orange) are indicated. The bars above the domain structures of both proteins mark the expressed fragments. (B) Gel filtration profiles of the yeast Seh1-Nup85 complex. The Seh1-Nup85 complex was injected at the indicated protein concentrations. The predicted elution positions for heter-tetrameric and hetero-octameric Seh1-Nup85 complexes are shown and have been determined using molecular weight standards.
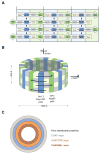
Figure 7. Model for the Architecture of the Symmetric NPC Core
(A, B) Eight Heptamers are circumferentially arranged in a head-to-tail fashion in four stacked rings. The Sec13-Nup145C and Seh1-Nup85 hetero-octamers serve as vertical poles connecting the four rings, thereby forming a scaffold. The poles are connected through their interaction with the remaining nups of the heptameric complex. Based on the two-fold axes of symmetry in the Sec13-Nup145C hetero-octamer, the heptameric complex rings are stacked with opposite directionality. (C) The symmetric core of the NPC is schematically represented as a series of concentric cylinders as discussed in the text. Each of the four envisaged cylinders would contain the principal mass of the following proteins: integral pore membrane proteins (Pom152, Pom34, and Ndc1), coat nups (Seh1, Nup85, Nup120, Sec13, Nup145C, Nup84, and Nup133), adapter nups (Nic96, Nup192, Nup188, Nup157, and Nup170), and the channel nups (Nsp1, Nup49, and Nup57).
Similar articles
- Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn SG, Schwartz TU. Brohawn SG, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1173-7. doi: 10.1038/nsmb.1713. Epub 2009 Oct 25. Nat Struct Mol Biol. 2009. PMID: 19855394 Free PMC article. - Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Liu X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16498-503. doi: 10.1073/pnas.1214557109. Epub 2012 Sep 26. Proc Natl Acad Sci U S A. 2012. PMID: 23019579 Free PMC article. - Structure of a trimeric nucleoporin complex reveals alternate oligomerization states.
Nagy V, Hsia KC, Debler EW, Kampmann M, Davenport AM, Blobel G, Hoelz A. Nagy V, et al. Proc Natl Acad Sci U S A. 2009 Oct 20;106(42):17693-8. doi: 10.1073/pnas.0909373106. Epub 2009 Oct 1. Proc Natl Acad Sci U S A. 2009. PMID: 19805193 Free PMC article. - The structure of the nuclear pore complex.
Hoelz A, Debler EW, Blobel G. Hoelz A, et al. Annu Rev Biochem. 2011;80:613-43. doi: 10.1146/annurev-biochem-060109-151030. Annu Rev Biochem. 2011. PMID: 21495847 Review. - Toward the molecular details of the nuclear pore complex.
Panté N, Aebi U. Panté N, et al. J Struct Biol. 1994 Nov-Dec;113(3):179-89. doi: 10.1006/jsbi.1994.1052. J Struct Biol. 1994. PMID: 7537513 Review. No abstract available.
Cited by
- Holes in the Nuclear Membrane as an Illustration of Gaps in the Understanding of the Biology by Biologists.
Kuvichkin V. Kuvichkin V. J Membr Biol. 2015 Aug;248(4):741-4. doi: 10.1007/s00232-015-9786-9. Epub 2015 Mar 11. J Membr Biol. 2015. PMID: 25758231 Review. - Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn SG, Schwartz TU. Brohawn SG, et al. Nat Struct Mol Biol. 2009 Nov;16(11):1173-7. doi: 10.1038/nsmb.1713. Epub 2009 Oct 25. Nat Struct Mol Biol. 2009. PMID: 19855394 Free PMC article. - Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats.
Lee C, Goldberg J. Lee C, et al. Cell. 2010 Jul 9;142(1):123-32. doi: 10.1016/j.cell.2010.05.030. Epub 2010 Jun 24. Cell. 2010. PMID: 20579721 Free PMC article. - Architecture of the linker-scaffold in the nuclear pore.
Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, Tomaleri GP, Schaus L, Hoelz A. Petrovic S, et al. Science. 2022 Jun 10;376(6598):eabm9798. doi: 10.1126/science.abm9798. Epub 2022 Jun 10. Science. 2022. PMID: 35679425 Free PMC article. - Structural evidence for common ancestry of the nuclear pore complex and vesicle coats.
Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Brohawn SG, et al. Science. 2008 Nov 28;322(5906):1369-73. doi: 10.1126/science.1165886. Epub 2008 Oct 30. Science. 2008. PMID: 18974315 Free PMC article.
References
- Allen NP, Huang L, Burlingame A, Rexach M. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem. 2001;276:29268–29274. - PubMed
- Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. - PubMed
- Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases