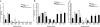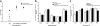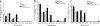Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract - PubMed (original) (raw)
Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract
Melissa Barman et al. Infect Immun. 2008 Mar.
Abstract
The commensal microbiota protects the murine host from enteric pathogens. Nevertheless, specific pathogens are able to colonize the intestinal tract and invade, despite the presence of an intact biota. Possibly, effective pathogens disrupt the indigenous microbiota, either directly through pathogen-commensal interaction, indirectly via the host mucosal immune response to the pathogen, or by a combination of these factors. This study investigates the effect of peroral Salmonella enterica serovar Typhimurium infection on the intestinal microbiota. Since the majority of the intestinal microbiota cannot be cultured by conventional techniques, molecular approaches using 16S rRNA sequences were applied. Several major bacterial groups were assayed using quantitative PCR. Administration of either the 50% lethal dose (LD(50)) or 10x LD(50) of Salmonella enterica serovar Typhimurium caused changes in the microbiota throughout the intestinal tract over the time course of infection. A 95% decrease in total bacterial number was noted in the cecum and large intestine with 10x LD(50) S. enterica serovar Typhimurium challenge at 7 days postinfection, concurrent with gross evidence of diarrhea. In addition, alterations in microbiota composition preceded the onset of diarrhea, suggesting the involvement of pathogen-commensal interactions and/or host responses unrelated to diarrhea. Microbiota alterations were not permanent and reverted to the microbiota of uninfected mice by 1 month postinfection. Infection with a Salmonella pathogenicity island 1 (SPI1) mutant did not result in microbiota alterations, while SPI2 mutant infections triggered partial changes. Neither mutant was capable of prolonged colonization or induction of mucosal inflammation. These data suggest that several Salmonella virulence factors, particularly those involved in the local mucosal host response, are required for disruption of the intestinal ecosystem.
Figures
FIG. 1.
Validation of qPCR for quantification of Salmonella and persistent colonization of the murine intestinal tract by Salmonella. (a) LB broth was coinoculated with S. enterica serovar Typhimurium and E. coli, which were grown at 37°C. An aliquot of culture was dilution plated on LB (to enumerate total bacteria) or SS (to enumerate Salmonella) agar. Total bacterial genomic DNA was isolated from an aliquot of culture and analyzed by qPCR to enumerate total bacteria and Salmonella. Black squares denote log numbers of bacteria determined by qPCR. Black triangles denote log numbers of bacteria determined by selective culture. (b) Mice were orally inoculated with either 108 CFU S. enterica serovar Typhimurium or buffer alone and then sacrificed at 3 or 7 days postinoculation. The intestinal tract was removed and divided. Bacterial genomic DNA was isolated from the DSI, cecum, and LI and analyzed by qPCR for quantification of Salmonella. No Salmonella cells were seen in control mice. Hatched bars represent infected mice at 3 days postinoculation. Black bars represent infected mice at 7 days postinoculation.
FIG. 2.
Characterization of enteric salmonellosis. Mice were orally inoculated with 108 CFU S. enterica serovar Typhimurium or buffer alone and then sacrificed after either 3 or 7 days. The mouse terminal ileum was removed and fixed in Carnoy's fixative. Hematoxylin and eosin staining was performed on terminal ileum sections from control (a) and _Salmonella_-infected (b and c) mice at 7 days postinfection. P, Peyer's patch; C, crypt epithelium; LP, lamina propria. Arrows indicate the locations of neutrophils. (d) The ceca of control and infected mice were removed and weighed for comparison at both 3 and 7 days postinfection. Black squares represent uninfected control mice. Black triangles represent _Salmonella_-infected mice. Bacterial genomic DNA was isolated from the DSI, ceca, and LI of control and infected mice at 3 (e) and 7 (f) days postinoculation and analyzed by qPCR for total bacteria. White bars represent uninfected controls. Black bars represent _Salmonella_-infected mice. *, P < 0.05 (Student's t test).
FIG. 3.
Quantitative analysis of intestinal microbiota 3 days after infection with 108 CFU S. enterica serovar Typhimurium. Mice were inoculated with 108 S. enterica serovar Typhimurium organisms and sacrificed after 3 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis measured the abundance of specific commensal bacterial groups in the DSI (a), cecum (b), and LI (c). White bars represent uninfected controls. Black bars represent _Salmonella_-infected mice. In the DSI, Salmonella infection did affect bacterial counts (P < 0.05), and the effect was not uniform across groups (_P_ < 0.05). The asterisk represents the post hoc _t_ test for the _Lactobacillus_ sp. group (_P_ < 0.005). _Salmonella_ infection did not appear to affect the cecum or LI (_P_ > 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
FIG. 4.
Quantitative analysis of intestinal microbiota 7 days after infection with 108 CFU S. enterica serovar Typhimurium. Mice were inoculated with 108 S. enterica serovar Typhimurium organisms and sacrificed after 7 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis measured the abundance of specific commensal bacterial groups in the DSI (a), cecum (b), and LI (c). White bars represent uninfected controls. Black bars represent _Salmonella_-infected mice. Salmonella infection did affect bacterial counts in the DSI (P < 0.0001), cecum (P < 0.0001), and LI (P < 0.0001), and the effects were not uniform across groups (P < 0.0001 for all segments). Asterisks represent the post hoc t test for the designated groups (P < 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
FIG. 5.
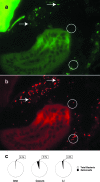
Localization of Salmonella in the terminal ileum. The mouse terminal ileum was removed from mice at 7 days post-peroral inoculation with 108 CFU S. enterica serovar Typhimurium and fixed in Carnoy's fixative. Three-micrometer sections were cut and analyzed by FISH to localize bacteria within the tissue section. Sections of small intestinal tissue were cohybridized with a combination of FAM-Sal and TR-Bact338 probes, which enabled visualization of the distribution of both Salmonella (a) and the total indigenous microbiota (b) within the small intestinal lumen. FAM-Sal specifically hybridizes to Salmonella (a), while TR-Bact338 shows total bacteria (b). Arrows show examples of Salmonella in each field. Circles illustrate examples of non-Salmonella indigenous bacteria. (c) qPCR was used to quantify total bacteria and Salmonella from the DSI, ceca, and LI of infected mice. The black section of each pie chart represents the percentage of the total microbiota comprised by Salmonella.
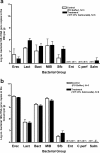
FIG. 6.
Impact of SPI1 and SPI2 Salmonella mutants on the intestinal microbiota. Mice were inoculated perorally with 108 CFU of S. enterica serovar Typhimurium TK93 (SPI1 mutant) or 5SAT (SPI2 mutant) and were sacrificed after 3 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. (a) Cecal weights were obtained and compared between control and infected mice. Black squares represent uninfected control mice. Black triangles represent _Salmonella_-infected mice. (b) Bacterial genomic DNA was isolated from the DSI of mice infected with the SPI1 mutant (b) and the SPI2 mutant (c), and qPCR analysis determining the abundance of specific commensal groups was performed. White bars represent uninfected controls. Black bars represent _Salmonella_-infected mice. The asterisk represents the post hoc t test for the designated group (P < 0.05). Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria.
FIG. 7.
Quantitative analysis of the intestinal microbiota after 7 days of infection with 107 CFU S. enterica serovar Typhimurium. Mice were inoculated perorally with 107 CFU S. enterica serovar Typhimurium and sacrificed after 7 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis was performed to determine the abundance of specific commensal bacterial groups in the DSI (a), cecum (b), and LI (c). White bars represent uninfected controls. Black bars represent _Salmonella_-infected mice. Salmonella infection did affect bacterial counts in the cecum (P < 0.005) and the LI (_P_ < 0.05), and the effects were not uniform across groups. Asterisks represent the post hoc _t_ test for the designated groups (_P_ < 0.005). _Salmonella_ infection did not appear to affect the bacterial counts in the small intestine (_P_ > 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
FIG. 8.
Quantitative analysis of the intestinal microbiota after intestinal clearance of S. enterica serovar Typhimurium. Mice were inoculated with 107 CFU S. enterica serovar Typhimurium and sacrificed after 30 days. The intestinal tract was removed and divided into the DSI, cecum, and LI. Bacterial genomic DNA was isolated from each segment, and qPCR analysis was performed to determine the abundance of specific commensal bacterial groups in the DSI (a) and cecum (b). White bars represent uninfected controls. Black bars represent _Salmonella_-infected mice. Salmonella infection did not appear to affect the bacterial numbers in any segment of the gut (P > 0.05). BT, below the detection threshold of qPCR. Erec, Eubacterium rectale/Clostridium coccoides; Lact, Lactobacillus sp.; Bact, Bacteroides sp.; MIB, mouse intestinal Bacteroides; Sfb, segmented filamentous bacteria; Ent, Enterobacteriaceae; C. perf, Clostridium perfringens; Salm, S. enterica serovar Typhimurium.
Comment in
- Intestinal microbiota are transiently altered during Salmonella-induced gastroenteritis.
Gibson DL, Vallance BA. Gibson DL, et al. Expert Rev Gastroenterol Hepatol. 2008 Aug;2(4):525-9. doi: 10.1586/17474124.2.4.525. Expert Rev Gastroenterol Hepatol. 2008. PMID: 19072400
References
- Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 3071915-1920. - PubMed
- Baker, G. C., J. J. Smith, and D. A. Cowan. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55541-555. - PubMed
- Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 712839-2858. - PMC - PubMed
- Bartosch, S., A. Fite, G. T. Macfarlane, and M. E. McMurdo. 2004. Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effect of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 703575-3581. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous