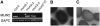The Arabidopsis MUM2 gene encodes a beta-galactosidase required for the production of seed coat mucilage with correct hydration properties - PubMed (original) (raw)
The Arabidopsis MUM2 gene encodes a beta-galactosidase required for the production of seed coat mucilage with correct hydration properties
Gillian H Dean et al. Plant Cell. 2007 Dec.
Abstract
Seed coat development in Arabidopsis thaliana involves a complex pathway where cells of the outer integument differentiate into a highly specialized cell type after fertilization. One aspect of this developmental process involves the secretion of a large amount of pectinaceous mucilage into the apoplast. When the mature seed coat is exposed to water, this mucilage expands to break the primary cell wall and encapsulate the seed. The mucilage-modified2 (mum2) mutant is characterized by a failure to extrude mucilage on hydration, although mucilage is produced as normal during development. The defect in mum2 appears to reside in the mucilage itself, as mucilage fails to expand even when the barrier of the primary cell wall is removed. We have cloned the MUM2 gene and expressed recombinant MUM2 protein, which has beta-galactosidase activity. Biochemical analysis of the mum2 mucilage reveals alterations in pectins that are consistent with a defect in beta-galactosidase activity, and we have demonstrated that MUM2 is localized to the cell wall. We propose that MUM2 is involved in modifying mucilage to allow it to expand upon hydration, establishing a link between the galactosyl side-chain structure of pectin and its physical properties.
Figures
Figure 1.
Cytological Analysis of the mum2 Mutant. (A) Mature wild-type seed stained with Ruthenium Red. A layer of stained mucilage is visible around the seed, and the columellae are clearly visible in the center of each cell, indicating that the outer tangential cell wall has ruptured. (B) Mature mum2 seed stained in Ruthenium Red. No extruded mucilage is visible, and the outer tangential cell walls have not ruptured. (C) Scanning electron microscopy of mature wild-type seed. Hexagonal epidermal cells with thickened radial walls and a central columella are visible. (D) Scanning electron microscopy of mature mum2 seed. The mutant seeds are indistinguishable from the wild type. (E) Wild-type seed coat epidermal cell at 4 DPA. Amyloplasts are visible; much of the cell volume is occupied by the vacuole. (F) Wild-type seed coat epidermal cell at 7 DPA. Mucilage secretion into the apoplast has begun, and the cytoplasmic column is visible. (G) Wild-type seed coat epidermal cell at 10 DPA. Mucilage secretion is complete, and columella formation has begun. (H) Wild-type seed coat epidermal cell at 13 DPA. Columella formation is complete. (I) Wild-type seed coat epidermal cell at 16 DPA. The epidermal cells of the seed coat have ruptured during the fixation process, and mucilage has been released. The columella and the radial cell walls are visible. (J) to (N) The development of the mum2 seed coat epidermal cells at 4 (J), 7 (K), 10 (L), and 13 DPA (M) is indistinguishable from the wild type. However, at 16 DPA (N), the mum2 seed coat does not rupture and the mucilage remains within the primary cell wall (cf. [I] with [N]). (O) and (P) Mature wild-type seed treated with EDTA (O) or Na2CO3 (P) and stained with Ruthenium Red. Mucilage has been released, and seeds have a similar appearance to those stained directly with Ruthenium Red (cf. with [A]). (Q) Mature mum2 seed treated with EDTA and stained with Ruthenium Red. Small amounts of mucilage are released from some cells. (R) Mature mum2 seed treated with Na2CO3 and stained with Ruthenium Red. A thin layer of poorly stained mucilage has been released. The columellae are visible, indicating that the outer tangential cell wall has ruptured (cf. [R] with [B]). (S) and (T) Sections (20 μm) of mature, unfixed, paraffin-embedded wild-type seed hydrated with Ruthenium Red (S) or Na2CO3 and then Ruthenium Red (T). The mucilage is extruded from the seed coat similar to intact seeds. (U) Section (20 μm) of mature, unfixed, paraffin-embedded mum2 seed hydrated with Ruthenium Red. The mucilage does not expand, despite being in direct contact with the aqueous dye solution. (V) Section (20 μm) of mature, unfixed, paraffin-embedded mum2 seed hydrated with Na2CO3 and then Ruthenium Red. The outer tangential cell wall has ruptured, and a thin layer of mucilage has been released, similar to intact seeds treated with Na2CO3. Bars = 100 μm in (A) to (D) and (O) to (V) and 10 μm in (E) to (N).
Figure 2.
Transcript Analysis in mum2 Alleles and Genomic Complementation of mum2 with At5g63800. (A) RT-PCR analyses on 7-DPA silique cDNA from, mum 2-2, mum2-5, and mum2-10 using _MUM2_-specific primers. Cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC) was used as a loading control. The larger product in mum2-1 caused by inefficient splicing is indicated by an arrow. (B) and (C) Ruthenium red–stained seeds. The mum2 plants transformed with pMUM2g show complementation of the mum2 phenotype as indicated by a restoration of wild-type mucilage extrusion (B). The mum2 plants transformed with empty pCAMBIA1200 retain a mum2 phenotype (C). Bars = 100 μm.
Figure 3.
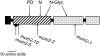
MUM2 Predicted Protein Structure. The predicted MUM2 protein is 718 amino acids long. The predicted signal peptide (solid box), the glycosyl hydrolase domain (striped box), and the galactose binding-like domains (dotted box) are indicated. Arrows indicate the locations of the ethyl methanesulfonate–induced mutations in mum2-1 (G513R) and mum2-2 (G238E), the deletion of amino acids 62 to 93 in mum2-5, and the position of the T-DNA between amino acids 99 and 100 in mum2-10. Arrows also indicate the positions of two predicted _N_-glycosylation sites (N-glyc), the predicted catalytic nucleophile, Glu-259 (N), and the predicted catalytic proton donor, Glu-188 (PD).
Figure 4.
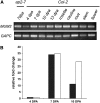
MUM2 Transcript Analysis. (A) RT-PCR analysis of MUM2 transcript in Arabidopsis tissues using gene-specific primers for MUM2. Cytosolic GAPC was used as a loading control. Silique RNA at 7 DPA from ap2 was used as a control for expression in seed and silique tissues other than the seed coat. (B) Real-time PCR analysis of MUM2 transcript during seed coat development. RNA was prepared from isolated seed coats at 4, 7, and 10 DPA, and MUM2 transcript levels were determined relative to GAPC using three technical replicates. Less than 3% variation in the Ct value was seen in the technical replicates for each transcript. Data are presented as relative fold change, where the 4-DPA transcript level was set at 1.0. Two biological replicates were performed, as indicated by black and white bars.
Figure 5.
Localization of MUM2-GFP Fusion Protein in Tobacco Epidermal Cells. (A) MUM2-GFP fusion protein is localized to the cell wall. Cell wall localization is characterized by a single line of fluorescence between the cells with no gaps at the corners of adjoining cells (arrow). Autofluorescence from chloroplasts in guard cells is also visible (c). To confirm this, the plasma membrane was counterstained with FM4-64 (inset, red). The pattern of staining, with the plasma membrane of adjoining cells separated by MUM2-GFP fluorescence, confirms that MUM2-GFP is located in the cell wall. (B) Localization of free GFP in the cytosol and nucleus (n). Cytosolic localization is characterized by the lack of fluorescence in the cell walls between cells (arrow) and is distinctly different to cell wall expression (cf. [A] with [B]). (C) MUM2-GFP coexpressed with AtRab1b(N121I) is retained in the ER. ER localization is characterized by fluorescence of the cortical network in tobacco epidermal cells (arrow). Autofluorescence from chloroplasts in guard cells is also visible (c). (D) Expression pattern of ER-retained GFP-HDEL. Again, the cortical network characteristic of ER is evident (arrow). This expression pattern is comparable with MUM2-GFP retained in the ER (cf. [C] with [D]). Bars = 30 μm.
Similar articles
- A naturally occurring mutation in an Arabidopsis accession affects a beta-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage.
Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. Macquet A, et al. Plant Cell. 2007 Dec;19(12):3990-4006. doi: 10.1105/tpc.107.050179. Epub 2007 Dec 28. Plant Cell. 2007. PMID: 18165330 Free PMC article. - A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats.
Rautengarten C, Usadel B, Neumetzler L, Hartmann J, Büssis D, Altmann T. Rautengarten C, et al. Plant J. 2008 May;54(3):466-80. doi: 10.1111/j.1365-313X.2008.03437.x. Epub 2008 Feb 7. Plant J. 2008. PMID: 18266922 - The transcriptional regulator LEUNIG_HOMOLOG regulates mucilage release from the Arabidopsis testa.
Walker M, Tehseen M, Doblin MS, Pettolino FA, Wilson SM, Bacic A, Golz JF. Walker M, et al. Plant Physiol. 2011 May;156(1):46-60. doi: 10.1104/pp.111.172692. Epub 2011 Mar 14. Plant Physiol. 2011. PMID: 21402796 Free PMC article. - Seed coat mucilage cells of Arabidopsis thaliana as a model for plant cell wall research.
Arsovski AA, Haughn GW, Western TL. Arsovski AA, et al. Plant Signal Behav. 2010 Jul;5(7):796-801. doi: 10.4161/psb.5.7.11773. Epub 2010 Jul 1. Plant Signal Behav. 2010. PMID: 20505351 Free PMC article. Review. - Regulation of seed coat mucilage production and modification in Arabidopsis.
Xu Y, Hu R, Li S. Xu Y, et al. Plant Sci. 2023 Mar;328:111591. doi: 10.1016/j.plantsci.2023.111591. Epub 2023 Jan 6. Plant Sci. 2023. PMID: 36623642 Review.
Cited by
- MUCILAGE-RELATED10 Produces Galactoglucomannan That Maintains Pectin and Cellulose Architecture in Arabidopsis Seed Mucilage.
Voiniciuc C, Schmidt MH, Berger A, Yang B, Ebert B, Scheller HV, North HM, Usadel B, Günl M. Voiniciuc C, et al. Plant Physiol. 2015 Sep;169(1):403-20. doi: 10.1104/pp.15.00851. Epub 2015 Jul 28. Plant Physiol. 2015. PMID: 26220953 Free PMC article. - Arabidopsis Seed Coat Mucilage is a Specialized Cell Wall that Can be Used as a Model for Genetic Analysis of Plant Cell Wall Structure and Function.
Haughn GW, Western TL. Haughn GW, et al. Front Plant Sci. 2012 Apr 3;3:64. doi: 10.3389/fpls.2012.00064. eCollection 2012. Front Plant Sci. 2012. PMID: 22645594 Free PMC article. - Spatiotemporal secretion of PEROXIDASE36 is required for seed coat mucilage extrusion in Arabidopsis.
Kunieda T, Shimada T, Kondo M, Nishimura M, Nishitani K, Hara-Nishimura I. Kunieda T, et al. Plant Cell. 2013 Apr;25(4):1355-67. doi: 10.1105/tpc.113.110072. Epub 2013 Apr 9. Plant Cell. 2013. PMID: 23572548 Free PMC article. - Emerging Functions for Cell Wall Polysaccharides Accumulated during Eudicot Seed Development.
Sechet J, Marion-Poll A, North HM. Sechet J, et al. Plants (Basel). 2018 Sep 29;7(4):81. doi: 10.3390/plants7040081. Plants (Basel). 2018. PMID: 30274256 Free PMC article. Review. - A naturally occurring mutation in an Arabidopsis accession affects a beta-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage.
Macquet A, Ralet MC, Loudet O, Kronenberger J, Mouille G, Marion-Poll A, North HM. Macquet A, et al. Plant Cell. 2007 Dec;19(12):3990-4006. doi: 10.1105/tpc.107.050179. Epub 2007 Dec 28. Plant Cell. 2007. PMID: 18165330 Free PMC article.
References
- Ahn, Y.O., Zheng, M., Bevan, D.R., Esen, A., Shiu, S.H., Benson, J., Peng, H.P., Miller, J.T., Cheng, C.L., Poulton, J.E., Shih, M.C. (2007). Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 35. Phytochemistry 68 1510–1520. - PubMed
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
- Balasubramaniam, S., Lee, H.C., Lazan, H., Othman, R., and Ali, Z.M. (2005). Purification and properties of a β-galactosidase from carambola fruit with significant activity towards cell wall polysaccharides. Phytochemistry 66 153–163. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases