Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres - PubMed (original) (raw)
Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres
Hernan Diego Folco et al. Science. 2008.
Abstract
Heterochromatin is defined by distinct posttranslational modifications on histones, such as methylation of histone H3 at lysine 9 (H3K9), which allows heterochromatin protein 1 (HP1)-related chromodomain proteins to bind. Heterochromatin is frequently found near CENP-A chromatin, which is the key determinant of kinetochore assembly. We have discovered that the RNA interference (RNAi)-directed heterochromatin flanking the central kinetochore domain at fission yeast centromeres is required to promote CENP-A(Cnp1) and kinetochore assembly over the central domain. The H3K9 methyltransferase Clr4 (Suv39); the ribonuclease Dicer, which cleaves heterochromatic double-stranded RNA to small interfering RNA (siRNA); Chp1, a component of the RNAi effector complex (RNA-induced initiation of transcriptional gene silencing; RITS); and Swi6 (HP1) are required to establish CENP-A(Cnp1) chromatin on naïve templates. Once assembled, CENP-A(Cnp1) chromatin is propagated by epigenetic means in the absence of heterochromatin. Thus, another, potentially conserved, role for centromeric RNAi-directed heterochromatin has been identified.
Figures
Fig. 1
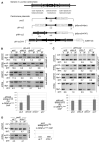
Testing roles for Clr4 in establishment of CENP-ACnp1 chromatin at centromeres. (A) Fission yeast centromere DNA and minichromosomes used (20). Association of CENP-ACnp1 and H3K9me2 with pH-cc2 (B), pHH-cc2 (C), or pcc2 (D) introduced into wild-type (wt) or clr4 cells by crossing (X) or transformation (T) is shown (see also fig. S1). Polymerase chain reaction (PCR) product positions on minichromosomes are indicated (A). fbp1 is the control noncentromeric locus. In CENP-ACnp1 ChIP, enrichment of pcc2 product was compared with input DNA (T) relative to fbp1. Enrichment of endogenous cc1/3 relative to fbp1 was also assessed. In H3K9me2 ChIP, enrichment at plasmid–outer repeats J2 (B) or K″K″ (C) products was compared with the PCR of input DNA relative to the fbp1 product. Enrichment of endogenous otr sequences was also assessed (B and C). Quantitative PCR confirms these results [lower panels (B) and (C) and right (D) and fig. S3] (20). Histograms show percent enrichment on plasmid relative to endogenous cc1/3.
Fig 2
Clr4 is required to establish, but not to maintain CENP-ACnp1 and kinetochore proteins on the large cen3 minichromosome. Association of CENP-ACnp1 and H3K9me2 in (A), and CENP-CCnp3 and Sim4 in (B) were determined by ChIP in wild-type and clr4 strains containing pH-icc3i-H introduced by crossing (X) or transformation (T). fbp1 or act1 are control noncentromeric loci. In CENP-ACnp1, CENP-CCnp3, and Sim4 ChIPs enrichment of plasmid cc3 product was compared with input DNA (T) relative to the act1 product. Positive control: Enrichment of endogenous centromeric cc1/3 was assessed. H3K9me2 ChIP: Enrichment of plasmid–outer repeat J3 product was compared with PCR of input DNA (T) relative to act1. Positive control: Enrichment of endogenous otr centromeric repeats was assessed.
Fig 3
clr4+ reintroduction triggers CENP-ACnp1 recruitment to minichromosome and minichromosome stability. (A) Association of H3K9me2 and CENP-ACnp1 with pH-icc3i-H in clr4 before and after clr4+ reintroduction (clr4+clr4). Primers and conditions as in Fig. 2. (B) Wild-type (wt) and clr4+clr4 cells containing pH-icc3i-H, are indistinguishable. Serial dilution assay on 1/10th adenine assesses minichromosome stability [red colonies (lost); white colonies (retained)] or full adenine ± 10 g/μl TBZ.
Fig 4
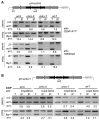
Swi6 and RNAi components are required for CENP-A establishment. CENP-ACnp1 and H3K9me2 ChIP on pHH-cc2 after direct transformation with plasmid DNA and selection in dcr1, chp1, swi6, and wild-type (wt) strains. Primers and conditions as in Fig 1C. Quantitative PCR confirms these results (lower panel and fig. S3) (20).
Similar articles
- Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres.
Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, Allshire RC. Kagansky A, et al. Science. 2009 Jun 26;324(5935):1716-9. doi: 10.1126/science.1172026. Science. 2009. PMID: 19556509 Free PMC article. - Heterochromatin protein 1 homologue Swi6 acts in concert with Ers1 to regulate RNAi-directed heterochromatin assembly.
Hayashi A, Ishida M, Kawaguchi R, Urano T, Murakami Y, Nakayama J. Hayashi A, et al. Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):6159-64. doi: 10.1073/pnas.1116972109. Epub 2012 Apr 2. Proc Natl Acad Sci U S A. 2012. PMID: 22474355 Free PMC article. - RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast.
Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF. Petrie VJ, et al. Mol Cell Biol. 2005 Mar;25(6):2331-46. doi: 10.1128/MCB.25.6.2331-2346.2005. Mol Cell Biol. 2005. PMID: 15743828 Free PMC article. - Studies on the mechanism of RNAi-dependent heterochromatin assembly.
Moazed D, Bühler M, Buker SM, Colmenares SU, Gerace EL, Gerber SA, Hong EJ, Motamedi MR, Verdel A, Villén J, Gygi SP. Moazed D, et al. Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review. - Heterochromatin tells CENP-A where to go.
Durand-Dubief M, Ekwall K. Durand-Dubief M, et al. Bioessays. 2008 Jun;30(6):526-9. doi: 10.1002/bies.20763. Bioessays. 2008. PMID: 18478529 Review.
Cited by
- Topoisomerase I is an evolutionarily conserved key regulator for satellite DNA transcription.
Teng Z, Yang L, Zhang Q, Chen Y, Wang X, Zheng Y, Tian A, Tian D, Lin Z, Deng WM, Liu H. Teng Z, et al. Nat Commun. 2024 Jun 17;15(1):5151. doi: 10.1038/s41467-024-49567-5. Nat Commun. 2024. PMID: 38886382 Free PMC article. - Topoisomerase I is an Evolutionarily Conserved Key Regulator for Satellite DNA Transcription.
Teng Z, Yang L, Zhang Q, Chen Y, Wang X, Zheng Y, Tian A, Tian D, Lin Z, Deng WM, Liu H. Teng Z, et al. bioRxiv [Preprint]. 2024 May 5:2024.05.03.592391. doi: 10.1101/2024.05.03.592391. bioRxiv. 2024. PMID: 38746280 Free PMC article. Updated. Preprint. - CFDP1 regulates the stability of pericentric heterochromatin thereby affecting RAN GTPase activity and mitotic spindle formation.
Gopinathan G, Xu Q, Luan X, Diekwisch TGH. Gopinathan G, et al. PLoS Biol. 2024 Apr 17;22(4):e3002574. doi: 10.1371/journal.pbio.3002574. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 38630655 Free PMC article. - Regional centromere configuration in the fungal pathogens of the Pneumocystis genus.
Cissé OH, Curran SJ, Folco HD, Liu Y, Bishop L, Wang H, Fischer ER, Davis AS, Combs C, Thapar S, Dekker JP, Grewal S, Cushion M, Ma L, Kovacs JA. Cissé OH, et al. mBio. 2024 Mar 13;15(3):e0318523. doi: 10.1128/mbio.03185-23. Epub 2024 Feb 21. mBio. 2024. PMID: 38380929 Free PMC article. - Calcineurin contributes to RNAi-mediated transgene silencing and small interfering RNA production in the human fungal pathogen Cryptococcus neoformans.
Yadav V, Mohan R, Sun S, Heitman J. Yadav V, et al. Genetics. 2024 Mar 6;226(3):iyae010. doi: 10.1093/genetics/iyae010. Genetics. 2024. PMID: 38279937
References
- Cleveland DW, Mao Y, Sullivan KF. Cell. 2003;112:407. - PubMed
- Sullivan BA, Blower MD, Karpen GH. Nat. Rev. Genet. 2001;2:584. - PubMed
- Morris CA, Moazed D. Cell. 2007;128:647. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials