The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses - PubMed (original) (raw)
The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses
Warawan Eiamphungporn et al. Mol Microbiol. 2008 Feb.
Abstract
The Bacillus subtilis extracytoplasmic function (ECF) sigma(M) factor is activated by cell envelope stress elicited by antibiotics, and by acid, heat, ethanol and superoxide stresses. Here, we have used several complementary approaches to identify genes controlled by sigma(M). In many cases, expression is only partially dependent on sigma(M) because of both overlapping promoter recognition with other ECF sigma factors and the presence of additional promoter elements. Genes regulated by sigma(M) have a characteristic pattern of induction in response to cell envelope-acting antibiotics as evidenced by hierarchical clustering analysis. sigma(M) also contributes to the expression of the Spx transcription factor and thereby indirectly regulates genes of the Spx regulon. Cell envelope stress responses also include regulons controlled by sigma(W), sigma(B) and several two-component regulatory systems (e.g. LiaRS, YycFG, BceRS). Activation of the sigma(M) regulon increases expression of proteins functioning in transcriptional control, cell wall synthesis and shape determination, cell division, DNA damage monitoring, recombinational repair and detoxification.
Figures
Figure 1
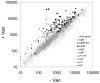
The vancomycin stimulon. Averaged signal intensity for each gene is plotted for cells grown in the absence of vancomycin (x-axis) or presence of vancomycin for 10 min. (y-axis). Genes induced by vancomycin were assigned to regulons as indicated in the inset. “sigM(new)” indicates those genes assigned to the σM regulon in this work.
Figure 2
σM activated genes assigned by ROMA. The σM-dependent increase in signal intensity is plotted vs. chromosome position. Note that the y-axis has been truncated so that genes with a large decrease in signal intensity are truncated at −500 pixel units (these typically result from strong σA-dependent promoters that are inhibited by addition of a large excess of σM). Gene symbols in larger font are assigned, based on the work reported herein, as at least partially σM-dependent in vivo.
Figure 3
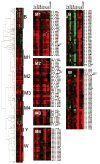
Hierarchical clustering analysis of gene expression in response to antibiotic stress. The antibiotic treatments included D-cycloserine (D-cyc), oxacillin (Oxa), amoxicillin (Amox), Ristocetin (Risto), gramicidin A (Gram), monensin (Mon), polymyxin B (Poly), Triton-X-114 (Tri), bacitracin (Bac), and vancomycin (Van) in both the wild-type and sigM mutant strains. Gene expression data was clustered based on expression level. Genes regulated by σB (B), σM (M1-M4), σY (Y), and σW (W) are highlighted. Red indicates induction, and green repression, under the specified treatment condition (see Experimental Procedures for details).
Figure 4
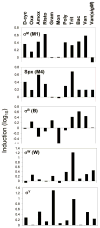
Characteristic induction profile of the σM, σW, σB, σY, and Spx regulons. The induction profiles (log scale) for the clusters highlighted in Fig. 3 are compared. See Figure 3 for abbreviations.
Figure 5
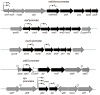
Gene organization for selected σM-regulated operons. Promoter sites are indicated by bent arrows with a subscript to indicate the relevant form(s) of holoenzyme. Genes in black are predicted to be cotranscribed based on gene organization and coexpression data (Sierro et al., 2007). The antisense oriented gene ydbP is shown in white. Transcription terminators are indicated by “lollipop” structures.
Figure 6
Dependence of selected promoters on σM, σX, and σW analysed with lacZ fusions. Reporter fusions were generated with cloned promoter regions integrated ectopically into SPβ. Cells were grown to mid-logarithmic phase and either induced or not with vancomycin for 10 min. prior to assay. Strains were assayed in the genetic backgrounds indicated in the inset where CU1065 represents wild-type and sigM indicates the sigM null mutant strain.
References
- Alba BM, Gross CA. Regulation of the Escherichia coliσE-dependent envelope stress response. Mol Microbiol. 2004;52:613–619. - PubMed
- Asai K, Yamaguchi H, Kang CM, Yoshida K, Fujita Y, Sadaie Y. DNA microarray analysis of Bacillus subtilis sigma factors of extracytoplasmic function family. FEMS Microbiol Lett. 2003;220:155–160. - PubMed
- Bauer R, Dicks LM. Mode of action of lipid II-targeting lantibiotics. Int J Food Microbiol. 2005;101:201–216. - PubMed
- Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases