Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification - PubMed (original) (raw)
Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification
Harm van Bakel et al. Nucleic Acids Res. 2008 Mar.
Abstract
Chromatin immunoprecipitation combined with DNA microarrays (ChIP-chip) is a powerful technique to detect in vivo protein-DNA interactions. Due to low yields, ChIP assays of transcription factors generally require amplification of immunoprecipitated genomic DNA. Here, we present an adapted linear amplification method that involves two rounds of T7 RNA polymerase amplification (double-T7). Using this we could successfully amplify as little as 0.4 ng of ChIP DNA to sufficient amounts for microarray analysis. In addition, we compared the double-T7 method to the ligation-mediated polymerase chain reaction (LM-PCR) method in a ChIP-chip of the yeast transcription factor Gsm1p. The double-T7 protocol showed lower noise levels and stronger binding signals compared to LM-PCR. Both LM-PCR and double-T7 identified strongly bound genomic regions, but the double-T7 method increased sensitivity and specificity to allow detection of weaker binding sites.
Figures
Figure 1.
Overview of the double-T7 RNA polymerase based linear amplification method. The steps in the T7 amplification protocol are illustrated for the top strand (black) of a double-stranded DNA template. The bottom strand (gray) is analogously amplified but omitted from this figure for clarity. After addition of poly(dT), an anchored T7-(dA)18 oligo is annealed and the overhangs are filled in using the Klenow fragment of DNA polymerase I. The resulting double-stranded DNA fragment contains a functional T7 promoter that drives the in vitro transcription reaction, generating multiple RNA copies of the DNA template. To increase the yield for low starting amounts of ChIP material, a second amplification round is performed after reverse transcription using random primers (RP = random primers).
Figure 2.
Yields obtained from amplification of different amounts of input DNA. An input DNA sample was obtained by reverse crosslinking chromatin extracts from a wild-type yeast culture, and used as starting material for a double-round T7 amplification. (A) Averaged RNA yields after two rounds of amplification are plotted as function of the amount of starting material used, which ranged from 0.4 to 2.5 ng. The concentration of T7 oligo added to the Klenow reactions was scaled proportionally with the amount of starting material, and set to 25 nM and 250 nM for the first and second round, respectively. Assays were performed in duplicate and error bars indicate the standard deviation. (B) Analysis of fragment size of amplified material after single round or double round of T7 amplification. One hundred and fifty nanograms of each sample was analyzed on an Agilent 2100 Bioanalyzer together with a RNA size marker. The fragment sizes of the RNA marker are indicated.
Figure 3.
Double-round T7 amplification results in reduced technical variation compared to LM-PCR. Histograms of averaged binding ratios of four hybridizations with double-T7 (A) or LM-PCR amplified samples (B) across all DNA microarray probes. (C) Overview plot of chromosome 7 showing locations of array probes (black) and genes (green). Graphs of binding ratios of input versus ChIP show the background and binding signals for double-round T7 amplification (double-T7) (top) and LM-PCR (bottom). (D) Enlargement of indicated regions in (C).
Figure 4.
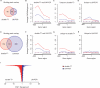
Comparison of significantly Gsm1p-bound regions after double-round T7 or LM-PCR amplification. The Gsm1p-bound regions identified after double-round T7 amplification (double-T7) and LM-PCR were compared after ANOVA analysis with high- (q < 0.05 and binding ratio>2) (A–D) or low-stringency (q < 0.05 and binding ratio > 1.5) cutoffs (E–H). Venn diagrams (A and E) indicate the degree of overlap between the binding peaks identified after double-T7 or LM-PCR amplification. Average binding profiles were determined for gene regions with significant binding on the ORF or 800 bp of promoter region as indicated (B–D, F–H). Averaged background binding profiles derived from randomly selected sets of genes of similar size are shown as dashed lines. (I) Direct comparison of the maximal peak height after double-T7 (red) or LM-PCR (blue) for all overlapping binding peaks identified in (E).
Figure 5.
Quantitative PCR validation of Gsm1p-bound regions identified after double-round T7 amplification and LM-PCR amplification. Quantitative PCR validation of Gsm1p-binding peaks identified by both double-round T7 amplification (double-T7) and LM-PCR (A), LM alone (B) or double-T7 alone (C). Binding to tested regions is represented as enrichment over the HMR silent mating-type locus. (D) Enlarged overview of Gsm1p-binding ratios on the indicated loci using either double-T7 (red) or LM-PCR (blue). Locations of array probes (black) and genes (green) are indicated.
Similar articles
- ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos.
Sandmann T, Jakobsen JS, Furlong EE. Sandmann T, et al. Nat Protoc. 2006;1(6):2839-55. doi: 10.1038/nprot.2006.383. Nat Protoc. 2006. PMID: 17406543 - ChIP-on-chip analysis of DNA topoisomerases.
Bermejo R, Katou YM, Shirahige K, Foiani M. Bermejo R, et al. Methods Mol Biol. 2009;582:103-18. doi: 10.1007/978-1-60761-340-4_9. Methods Mol Biol. 2009. PMID: 19763945 - Mapping the distribution of chromatin proteins by ChIP on chip.
Nègre N, Lavrov S, Hennetin J, Bellis M, Cavalli G. Nègre N, et al. Methods Enzymol. 2006;410:316-41. doi: 10.1016/S0076-6879(06)10015-4. Methods Enzymol. 2006. PMID: 16938558 Review. - Genomic analysis of protein-DNA interactions in bacteria: insights into transcription and chromosome organization.
Wade JT, Struhl K, Busby SJ, Grainger DC. Wade JT, et al. Mol Microbiol. 2007 Jul;65(1):21-6. doi: 10.1111/j.1365-2958.2007.05781.x. Mol Microbiol. 2007. PMID: 17581117 Review.
Cited by
- Single-tube linear DNA amplification for genome-wide studies using a few thousand cells.
Shankaranarayanan P, Mendoza-Parra MA, van Gool W, Trindade LM, Gronemeyer H. Shankaranarayanan P, et al. Nat Protoc. 2012 Jan 26;7(2):328-38. doi: 10.1038/nprot.2011.447. Nat Protoc. 2012. PMID: 22281868 - The organization of genome duplication is a critical determinant of the landscape of genome maintenance.
Gómez-Escoda B, Wu PJ. Gómez-Escoda B, et al. Genome Res. 2018 Aug;28(8):1179-1192. doi: 10.1101/gr.224527.117. Epub 2018 Jun 22. Genome Res. 2018. PMID: 29934426 Free PMC article. - Replication origin selection regulates the distribution of meiotic recombination.
Wu PY, Nurse P. Wu PY, et al. Mol Cell. 2014 Feb 20;53(4):655-62. doi: 10.1016/j.molcel.2014.01.022. Mol Cell. 2014. PMID: 24560273 Free PMC article. - HyCCAPP as a tool to characterize promoter DNA-protein interactions in Saccharomyces cerevisiae.
Guillen-Ahlers H, Rao PK, Levenstein ME, Kennedy-Darling J, Perumalla DS, Jadhav AY, Glenn JP, Ludwig-Kubinski A, Drigalenko E, Montoya MJ, Göring HH, Anderson CD, Scalf M, Gildersleeve HI, Cole R, Greene AM, Oduro AK, Lazarova K, Cesnik AJ, Barfknecht J, Cirillo LA, Gasch AP, Shortreed MR, Smith LM, Olivier M. Guillen-Ahlers H, et al. Genomics. 2016 Jun;107(6):267-73. doi: 10.1016/j.ygeno.2016.05.002. Epub 2016 May 13. Genomics. 2016. PMID: 27184763 Free PMC article. - MRE11-RAD50-NBS1 promotes Fanconi Anemia R-loop suppression at transcription-replication conflicts.
Chang EY, Tsai S, Aristizabal MJ, Wells JP, Coulombe Y, Busatto FF, Chan YA, Kumar A, Dan Zhu Y, Wang AY, Fournier LA, Hieter P, Kobor MS, Masson JY, Stirling PC. Chang EY, et al. Nat Commun. 2019 Sep 19;10(1):4265. doi: 10.1038/s41467-019-12271-w. Nat Commun. 2019. PMID: 31537797 Free PMC article.
References
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. - PubMed
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. - PubMed
- Hecht A, Strahl-Bolsinger S, Grunstein M. Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol. Biol. 1999;119:469–479. - PubMed
- Strutt H, Paro R. Mapping DNA target sites of chromatin proteins in vivo by formaldehyde crosslinking. Methods Mol. Biol. 1999;119:455–467. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases