Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids - PubMed (original) (raw)
Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids
Qun Pan et al. J Cell Mol Med. 2008 Jan-Feb.
Retraction in
- Retracted: Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids.
Pan Q, Luo X, Chegini N. Pan Q, et al. J Cell Mol Med. 2015 Oct;19(10):2512. doi: 10.1111/jcmm.12621. J Cell Mol Med. 2015. PMID: 26411639 Free PMC article.
Abstract
Given the emerging roles of microRNAs (miRNAs) as key regulator of mRNA stability we assessed their expression profile in paired myometrium and leiomyoma, their isolated smooth muscle cells (MSMC and LSMC), a spontaneously transformed leiomyoma smooth muscle cells (T-LSMC) and SK-LMS-1, a leiomyosarcoma cell line using microarray and real time PCR. Based on global normalization of expression values of 385 miRNAs and statistical analysis (anova), 91 miRNAs were expressed above the threshold levels in myometrium, with a progressive decline in numbers in leiomyomas, MSMC, LSMC, T-LSMC and SK-LMS-1 (P<0.05). We selected and validated the expression of miR-20a, miR-21, miR-26a, miR-18a, miR-206, miR-181a and miR-142-5p and found their differential expression in tissue and cell-specific manners (P<0.05). Treatments of MSMC and LSMC with 17beta estradiol and medroxyprogesterone acetate (10(-8)M), or ICI-182780 and RU-486 (10(-6)M) resulted in differential regulation of these miRNAs (P<0.05). In conclusion, the expression of a number of miRNAs in myometrium and leiomyoma with their progressive aberrant from normal MSMC into LSMC, transformed and cancerous stage, suggests that miRNAs and their regulation by ovarian steroids play a key role in pathogenesis of leiomyoma through gene expression stability.
Figures
1
Hierarchical cluster and tree-view analysis of differentially expressed miRNAs profile in paired myometrium (MY1, MY2, MY3 and MY4) and leiomyomas (LY1, LY2, LY3 and LY4) (Fig. 1A) and in paired myometrial smooth muscle cells (MSMC1 and MSMC2) and leiomyoma smooth muscle cells (LSMC1 and LSMC2) isolated and cultured from the above tissues, transformed leiomyoma cells (TLSMC-1 and T-LSMC-2) and leiomyosarcoma cell line (SLKMS1 and SKLMS2). Each column represents data from a single cohort with shades of red and green indicating up- or down-regulated miRNA according to the colour scheme shown below.miRNAs represented by rows were clustered according to their similarities in pattern of expression in each tissue. The dendrogram at the top of the image displays similarity of expression among these cohorts (see Table 1 and Table 2 for the list of differentially expressed miRNAs).
2
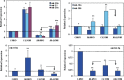
Bar graphs show real time PCR expression of miR-20a, miR-21 and miR-26a, miR-18a, miR-206, miR-181a and miR-142-5p in paired myometrium (Myo) and leiomyomas (LYOM) form African-Americans (AA) and Caucasians (C). The data is presented as relative expression following normalization and setting the expression of each miRNA independently in Caucasian myometrium arbitrarily as 1.Data represent mean ±standard error of three paired tissues from each ethnic group with asterisks *, ** and *** significantly different from C-Myo. P<0.05 was considered significant. Arrows indicates significant difference between the expression of these miRNAs in AA-Myo and AA-LYMO.
3
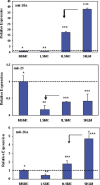
Bar graphs show real time PCR expression of miR-20a, miR-21 and miR-26a in myometrial and leiomyoma smooth muscle cells (MSMC and LSMC), spontaneously transformed LSMC (tLSMC) and leiomyosarcoma cell line, SKLM-1. The cells (1_106/well in 6-well plates) were cultured as described in materials and methods and their total RNA was isolated and subjected to real time PCR. The data is presented as relative expression following normalization and setting their expression level in MSMC arbitrarily as 1. Data represent mean ± standard error from three separate experiments, with asterisks ** and *** displaying significant difference with * with P<0.05 considered significant. Arrow indicates significant difference in the expression of these miRNAs between the cell types.
4
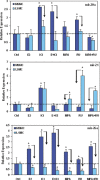
Bar graphs show the expression of miR-20a, miR-21 and miR-26a in myometrial and leiomyoma smooth muscle cells (MSMC and LSMC).The cells (1_106/well in 6-well plates) were cultured as described in materials and methods and following 24 hrs of treatment with 17b estradiol (E2), ICI-182780 (ICI), E2+ICI, medroxyprog-esterone acetate (MPA), RU-486 (RU) and MPA+RU, their total RNA was isolated and subjected to real time PCR. The data is presented as relative expression following normalization and setting their expression level in untreated MSMC arbitrarily as 1. Data represent mean ± standard error from three separate experiments. The asterisks * indicate statistical difference between the expression of these miRNAs in treated as compared to untreated controls (Ctrl), with arrows pointing out the difference in their expression between MSMC and LSMC. A probability level of P<0.05 was considered significant.
Comment in
- Findings of Research Misconduct.
[No authors listed] [No authors listed] Fed Regist. 2017 Aug 3;82(148):36150-36151. Fed Regist. 2017. PMID: 28857091 Free PMC article. No abstract available.
Similar articles
- microRNA 21: response to hormonal therapies and regulatory function in leiomyoma, transformed leiomyoma and leiomyosarcoma cells.
Pan Q, Luo X, Chegini N. Pan Q, et al. Mol Hum Reprod. 2010 Mar;16(3):215-27. doi: 10.1093/molehr/gap093. Epub 2009 Nov 11. Mol Hum Reprod. 2010. PMID: 19906824 Free PMC article. Retracted. - The expression of Abl interactor 2 in leiomyoma and myometrium and regulation by GnRH analogue and transforming growth factor-beta.
Luo X, Levens E, Williams RS, Chegini N. Luo X, et al. Hum Reprod. 2006 Jun;21(6):1380-6. doi: 10.1093/humrep/del011. Epub 2006 Feb 17. Hum Reprod. 2006. PMID: 16488906 - Regulation of transforming growth factor-beta1 expression by granulocyte macrophage-colony-stimulating factor in leiomyoma and myometrial smooth muscle cells.
Chegini N, Tang XM, Ma C. Chegini N, et al. J Clin Endocrinol Metab. 1999 Nov;84(11):4138-43. doi: 10.1210/jcem.84.11.6147. J Clin Endocrinol Metab. 1999. PMID: 10566662 - Non-coding RNAs in Uterine Development, Function and Disease.
Nothnick WB. Nothnick WB. Adv Exp Med Biol. 2016;886:171-189. doi: 10.1007/978-94-017-7417-8_9. Adv Exp Med Biol. 2016. PMID: 26659492 Free PMC article. Review. - The Usefulness of Immunohistochemistry in the Differential Diagnosis of Lesions Originating from the Myometrium.
Rubisz P, Ciebiera M, Hirnle L, Zgliczyńska M, Łoziński T, Dzięgiel P, Kobierzycki C. Rubisz P, et al. Int J Mol Sci. 2019 Mar 6;20(5):1136. doi: 10.3390/ijms20051136. Int J Mol Sci. 2019. PMID: 30845657 Free PMC article. Review.
Cited by
- MicroRNA expression and regulation in human ovarian carcinoma cells by luteinizing hormone.
Cui J, Eldredge JB, Xu Y, Puett D. Cui J, et al. PLoS One. 2011;6(7):e21730. doi: 10.1371/journal.pone.0021730. Epub 2011 Jul 12. PLoS One. 2011. PMID: 21765906 Free PMC article. - An emerging role for microRNAs in sexually dimorphic neurobiological systems.
Pak TR, Rao YS, Prins SA, Mott NN. Pak TR, et al. Pflugers Arch. 2013 May;465(5):655-67. doi: 10.1007/s00424-013-1227-y. Epub 2013 Feb 9. Pflugers Arch. 2013. PMID: 23397171 Free PMC article. Review. - Role of microRNA-21 and programmed cell death 4 in the pathogenesis of human uterine leiomyomas.
Fitzgerald JB, Chennathukuzhi V, Koohestani F, Nowak RA, Christenson LK. Fitzgerald JB, et al. Fertil Steril. 2012 Sep;98(3):726-734.e2. doi: 10.1016/j.fertnstert.2012.05.040. Epub 2012 Jun 22. Fertil Steril. 2012. PMID: 22728051 Free PMC article. - An induction of microRNA, miR-7 through estrogen treatment in breast carcinoma.
Masuda M, Miki Y, Hata S, Takagi K, Sakurai M, Ono K, Suzuki K, Yang Y, Abe E, Hirakawa H, Ishida T, Suzuki T, Ohuchi N, Sasano H. Masuda M, et al. J Transl Med. 2012 Sep 19;10 Suppl 1(Suppl 1):S2. doi: 10.1186/1479-5876-10-s1-s2. J Transl Med. 2012. PMID: 23227519 Free PMC article. - Sex differences in microRNA regulation of gene expression: no smoke, just miRs.
Morgan CP, Bale TL. Morgan CP, et al. Biol Sex Differ. 2012 Sep 26;3(1):22. doi: 10.1186/2042-6410-3-22. Biol Sex Differ. 2012. PMID: 23009289 Free PMC article.
References
- Varol N, Healey M, Tang P, Sheehan P, Maher P, Hill D. Ten-year review of hysterectomy morbidity and mortality: Can we change direction? Aust N Z J Obstet Gynaecol. 2001;41:295–302. - PubMed
- Al Hendy A, Salama SA. Catechol-o-methyltransferase polymorphism is associated with increased uterine leiomyoma risk in different ethnic groups. J Soc Gynecol Investig. 2006;13:136–44. - PubMed
- Denschlag D, Bentz EK, Hefler L, Pietrowski D, Zeillinger R, Tempfer C, Tong D. Genotype distribution of estrogen receptor-alpha, catechol-o-methyl-transferase, and cytochrome P450 17 gene polymorphisms in caucasian women with uterine leiomy-omas. Fertil Steril. 2006;85:462–7. - PubMed
- Villanova FE, Andrade PM, Otsuka AY, Gomes MT, Leal ES, Castro RA, Girao MJ, Nishimura E, Baracat EC, Silva ID. Estrogen receptor alpha polymorphism and susceptibility to uterine leiomyoma. Steroids. 2006;71:960–5. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical