Endogenous human microRNAs that suppress breast cancer metastasis - PubMed (original) (raw)
Endogenous human microRNAs that suppress breast cancer metastasis
Sohail F Tavazoie et al. Nature. 2008.
Abstract
A search for general regulators of cancer metastasis has yielded a set of microRNAs for which expression is specifically lost as human breast cancer cells develop metastatic potential. Here we show that restoring the expression of these microRNAs in malignant cells suppresses lung and bone metastasis by human cancer cells in vivo. Of these microRNAs, miR-126 restoration reduces overall tumour growth and proliferation, whereas miR-335 inhibits metastatic cell invasion. miR-335 regulates a set of genes whose collective expression in a large cohort of human tumours is associated with risk of distal metastasis. miR-335 suppresses metastasis and migration through targeting of the progenitor cell transcription factor SOX4 and extracellular matrix component tenascin C. Expression of miR-126 and miR-335 is lost in the majority of primary breast tumours from patients who relapse, and the loss of expression of either microRNA is associated with poor distal metastasis-free survival. miR-335 and miR-126 are thus identified as metastasis suppressor microRNAs in human breast cancer.
Figures
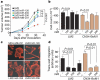
Figure 1. Systematic identification of miRNAs that suppress lung and bone metastasis in multiple human breast cancer cell derivatives
a, Hierarchical clustering of normalized miRNA expression levels for the 20 miRNAs that displayed the highest coefficients of variation clusters the MDA-MB-231 derivatives into three groups comprised of the bone metastatic BM2 lines (2287 and 1833), the lung metastatic LM2 lines (4180, 4173, 4175, and 4142) and the parental MDA lines. The heatmap highlights a set of miRNAs whose expression decreases across all lung and bone metastatic derivatives (red bracket along the right). The scale bar across the bottom depicts standard deviation change from the mean. b, Bioluminescence imaging of lung metastasis by lung metastatic breast cancer cells with restored expression of specific miRNAs. 1 × 104 LM2 cells expressing individual miRNAs or the control hairpin, as well as the parental MDA-MB-231 cells, were inoculated intravenously into immunodeficient mice. Shown are representative mice corresponding to the LM2 set (top) and the MDA-MB-231 set (bottom) at day 50. Lung colonization was measured by bioluminescence and quantified. n = 5; error bars represent s.e.m.; asterisk, P < 0.05. c, Human-vimentin-stained images of representative lungs that emitted the median luciferase signal for each cohort. Lungs were extracted at 8 weeks after xenografting and representative images of sections were obtained at whole-field magnification. d,2 × 104 BM2 cells expressing individual miRNAs or the control hairpin were inoculated into the arterial circulation via intracardiac injection of athymic mice. Bone metastasis was measured by bioluminescence and quantified as the normalized hindlimb photon flux. Shown are representative mice corresponding to marked data points. n = 6-10; horizontal line represents median signal for each cohort; _P_-values based on a one-tailed rank-sum test. e, Representative whole-field magnification images of haematoxylin- and eosin-stained femurs extracted from representative mice 7 weeks after intraventricular injection of indicated cancer cells. f, 2 × 105 primary human cancer derivative CN34-LM1 cells expressing individual miRNAs or the control vector were inoculated intravenously into immunodeficient mice. Lung colonization was measured by bioluminescence, quantified and normalized. n = 8; error bars represent s.e.m.; _P_-values based on a one-sided rank-sum test. g, 2 × 105 primary human cancer derivative CN34-BoM1 cells expressing individual miRNAs or the control vector were inoculated into the arterial circulation via intracardiac injection of immunodeficient mice. Bone metastasis was measured by bioluminescence, quantified and normalized. n = 9-10; error bars represent s.e.m.; _P_-values based on a one-tailed rank-sum test.
Figure 2. miR-126 suppresses overall tumour growth and proliferation whereas miR-335 and miR-206 regulate migration and morphology
a, 5 × 105 LM2 cells expressing individual miRNAs or the control hairpin, as well as the parental MDA-MB-231 cells, were injected into the mammary fat pads of immunodeficient mice and tumour volumes were measured over time. n = 5 (MDA-MB-231, LM2/miR-126, LM2/miR-206) and n = 10 (LM2, LM2/miR-335); error bars indicate s.e.m.; _P_-values based on a one-sided Student's _t_-test at day 32. b, 5 × 104 LM2 or primary breast cancer line CN34-BoM1 expressing miR-126, miR-335 or miR-206 and control cells were seeded in triplicate and viable cells were counted at 5 days after seeding. n = 3; error bars represent s.e.m.; _P_-values obtained using a one-sided Student's _t_-test. c, Parental MDA-MB-231 cells and lung metastatic LM2 cells expressing miR-126, miR-335 or miR-206 were seeded onto glass slides. Cells were stained with the actin marker phalloidin and confocal images were obtained. d, 2.5 × 104 LM2 and CN34-BoM1 cells were transduced with the indicated miRNAs or a control hairpin, and trans-well migration was assessed. Images of cells that had migrated through trans-well inserts were obtained and analysed in automated fashion using Metamorph software. n = 3; error bars represent s.e.m.; _P_-values obtained using a one-sided Student's _t_-test.
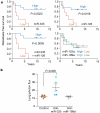
Figure 3. Clinical association of miR-335 and miR-126 with metastasis-free survival
a, miR-335, miR-126, miR-122a and miR-199a expression was assessed in a set of 20 primary breast tumour samples through qRT-PCR. Kaplan-Meier curves depict metastasis-free survival of patients whose primary tumours contained low or high levels of the indicated miRNAs. _P_-values were obtained using a log-rank test. b, The parental MDA-MB-231 cells were transfected with antagomirs targeting endogenous miR-335, miR-199a* or a control antagomir. Four days after transfection, 1 × 104 LM2 cells from each cohort were inoculated intravenously into immunodeficient mice. Lung colonization was measured by bioluminescence at day 35 and quantified. n = 5; error bars represent s.e.m.; _P_-values based on a one-sided rank-sum test.
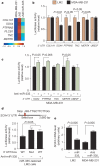
Figure 4. A miR-335-regulated gene set includes SOX4 as a miR-335 direct target
a, The miR-335 metastasis signature consists of genes downregulated by miR-335 and are also overexpressed in bone as well as lung metastatic MDA-MB-231 derivatives. The heatmap depicts the variance-normalized expression values for each gene averaged across two MDA-MB-231 samples, four LM2 samples and two LM2/miR-335 samples. The scale bar depicts standard deviation change from the mean for the expression value of each gene. b, UTR reporter assays of miR-335 metastasis genes in LM2 and MDA-MB-231 cells. Reporter constructs consisting of the luciferase sequence fused to the 3′ UTRs of the miR-335 metastasis genes as well as the control gene UBE2F were transfected into the LM2 and parental MDA-MB-231 cell lines. Luciferase activity of cells was assayed at 32 h after transfection and the values were normalized to the LM2 cell line. n = 3; error bars represent s.e.m.; _P_-values were obtained using a one-sided Student's _t_-test. c, Luciferase activity of MDA-MB-231 cells co-transfected with 39 UTR reporter constructs with, or without, miR-335 antagomir was assayed at 32 h after transfection and normalized to control cells. n = 3; error bars represent s.e.m.; _P_-values obtained using a one-sided Student's _t_-test. d, Schematic diagram depicts seed sequence (red box) in SOX4 UTR. The 60 bp containing this sequence or the miR-335 seed target site mutant were subjected to UTR reporter assays in LM2 cells with restored miR-335 expression. Luciferase activity of cells was also assayed in the presence or absence of the miR-335 antagomir. n = 3; error bars represent s.e.m.; _P_-values obtained using a one-sided Student's _t_-test. e, SOX4 expression in LM2 and MDA-MB-231 cells was obtained through real time qRT-PCR in cells expressing miR-335 or transfected with the miR-335 antagomir. n = 3; error bars represent s.e.m.; _P_-values derived using a one-sided Student's _t_-test.
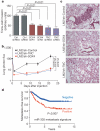
Figure 5. miR-335 regulates metastasis and invasion through suppression of SOX4 and TNC
a, 5.0 × 104 LM2 cells were transduced with short hairpin control vector, either of two shRNAs targeting SOX4, a hairpin targeting TNC, or an siRNA targeting TNC, and invasion of cells through a trans-well matrigel-coated membrane insert was quantified. n = 6; error bars represent s.e.m.; _P_-values based on a one-sided Student's _t_-test. b, 2 × 105 LM2 cells transduced with either a control short hairpin, a short hairpin targeting SOX4 or one targeting TNC were inoculated intravenously into immunodeficient mice. Lung colonization was measured by bioluminescence and quantified. n = 7; error bars represent s.e.m.; _P_-values based on a one-tailed rank-sum test. c, Haematoxylin-and-eosin-stained images of representative lungs that were extracted at 4 weeks after cell inoculation. Representative images were obtained at 10× magnification. d, Kaplan-Meier curves for the combined Memorial Sloan Kettering and Erasmus Medical Center breast tumour cohorts (368 tumours) depicting metastasis-free survival of patients whose primary tumours expressed the miR-335 six-gene signature (positive) and those that did not (negative). n = 368; _P_-value based on the Mantel-Cox log-rank test.
Similar articles
- MicroRNA-340 inhibits the migration, invasion, and metastasis of breast cancer cells by targeting Wnt pathway.
Mohammadi-Yeganeh S, Paryan M, Arefian E, Vasei M, Ghanbarian H, Mahdian R, Karimipoor M, Soleimani M. Mohammadi-Yeganeh S, et al. Tumour Biol. 2016 Jul;37(7):8993-9000. doi: 10.1007/s13277-015-4513-9. Epub 2016 Jan 12. Tumour Biol. 2016. PMID: 26758430 - Tumour invasion and metastasis initiated by microRNA-10b in breast cancer.
Ma L, Teruya-Feldstein J, Weinberg RA. Ma L, et al. Nature. 2007 Oct 11;449(7163):682-8. doi: 10.1038/nature06174. Epub 2007 Sep 26. Nature. 2007. PMID: 17898713 - Breast cancer metastasis: a microRNA story.
Negrini M, Calin GA. Negrini M, et al. Breast Cancer Res. 2008;10(2):203. doi: 10.1186/bcr1867. Epub 2008 Mar 26. Breast Cancer Res. 2008. PMID: 18373886 Free PMC article. - A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells.
Png KJ, Halberg N, Yoshida M, Tavazoie SF. Png KJ, et al. Nature. 2011 Dec 14;481(7380):190-4. doi: 10.1038/nature10661. Nature. 2011. PMID: 22170610 - Metastamir: the field of metastasis-regulatory microRNA is spreading.
Hurst DR, Edmonds MD, Welch DR. Hurst DR, et al. Cancer Res. 2009 Oct 1;69(19):7495-8. doi: 10.1158/0008-5472.CAN-09-2111. Epub 2009 Sep 22. Cancer Res. 2009. PMID: 19773429 Free PMC article. Review.
Cited by
- Clinical and prognostic association of transcription factor SOX4 in gastric cancer.
Fang CL, Hseu YC, Lin YF, Hung ST, Tai C, Uen YH, Lin KY. Fang CL, et al. PLoS One. 2012;7(12):e52804. doi: 10.1371/journal.pone.0052804. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23285187 Free PMC article. - Association of germline microRNA SNPs in pre-miRNA flanking region and breast cancer risk and survival: the Carolina Breast Cancer Study.
Bensen JT, Tse CK, Nyante SJ, Barnholtz-Sloan JS, Cole SR, Millikan RC. Bensen JT, et al. Cancer Causes Control. 2013 Jun;24(6):1099-109. doi: 10.1007/s10552-013-0187-z. Epub 2013 Mar 23. Cancer Causes Control. 2013. PMID: 23526039 Free PMC article. - MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review.
Iorio MV, Croce CM. Iorio MV, et al. EMBO Mol Med. 2012 Mar;4(3):143-59. doi: 10.1002/emmm.201100209. Epub 2012 Feb 20. EMBO Mol Med. 2012. PMID: 22351564 Free PMC article. Review. - Interplay between β1-integrin and Rho signaling regulates differential scattering and motility of pancreatic cancer cells by snail and Slug proteins.
Shields MA, Krantz SB, Bentrem DJ, Dangi-Garimella S, Munshi HG. Shields MA, et al. J Biol Chem. 2012 Feb 24;287(9):6218-29. doi: 10.1074/jbc.M111.308940. Epub 2012 Jan 9. J Biol Chem. 2012. PMID: 22232555 Free PMC article. - Do microRNA 96, 145 and 221 expressions really aid in the prognosis of prostate carcinoma?
Kang SG, Ha YR, Kim SJ, Kang SH, Park HS, Lee JG, Cheon J, Kim CH. Kang SG, et al. Asian J Androl. 2012 Sep;14(5):752-7. doi: 10.1038/aja.2012.68. Epub 2012 Aug 6. Asian J Androl. 2012. PMID: 22864280 Free PMC article.
References
- Fidler IJ. The pathogenesis of cancer metastasis: the `seed and soil' hypothesis revisited. Nature Rev. Cancer. 2003;3:453–458. - PubMed
- Weigelt B, Peterse JL, van't Veer LJ. Breast cancer metastasis: markers and models. Nature Rev. Cancer. 2005;5:591–602. - PubMed
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. - PubMed
- Nevins JR, Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nature Rev. Genet. 2007;8:601–609. - PubMed
- Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nature Rev. Genet. 2007;8:341–352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials