From individual Wnt pathways towards a Wnt signalling network - PubMed (original) (raw)
Review
From individual Wnt pathways towards a Wnt signalling network
Hans A Kestler et al. Philos Trans R Soc Lond B Biol Sci. 2008.
Abstract
Wnt proteins play important roles during vertebrate and invertebrate development. They obviously have the ability to activate different intracellular signalling pathways. Based on the characteristic intracellular mediators used, these are commonly described as the Wnt/beta-catenin, the Wnt/calcium and the Wnt/Jun N-terminal kinase pathways (also called planar cell polarity pathway). In the past, these different signalling events were mainly described as individual and independent signalling branches. Here, we discuss the possibility that Wnt proteins activate a complex intracellular signalling network rather than individual pathways and suggest a graph representation of this network. Furthermore, we discuss different ways of how to predict the specific outcome of an activation of this network in a particular cell type, which will require the use of mathematical models. We point out that the use of deterministic approaches via the application of differential equations is suitable to model only small aspects of the whole network and that more qualitative approaches are possibly a suitable starting point for the prediction of the global behaviour of such large protein interaction networks.
Figures
Figure 1
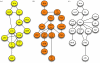
The classical view of three independent Wnt signalling pathways. Graph representations are given, including the core components of the (a) canonical Wnt/β-catenin pathway (yellow), (b) the Wnt/calcium pathway (orange) and (c) the Wnt/JNK pathway (white, also called planar cell polarity pathway). Edges indicate direct protein–protein interactions or may relate to an epistatic relationship between two components within a signalling pathway.
Figure 2
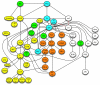
The integrative approach of analysing Wnt signalling results in a Wnt signalling network. Several components have been linked to all signalling branches such as Wnt, Fz, heterotrimeric G-proteins, dsh or PKC (indicated in light blue); others were linked to at least two branches such as CKIϵ, diversin, dkk, cdc 42 or CKIα (indicated in green). Please note that this network does not represent all interactions identified in Wnt signalling so far for reasons of clarity. Most of the edges represent direct interactions of entities indicated and some of them however represent the epistatic relationship of these components within the network as discussed in the main text. Dashed lines for the upstream molecules of yes indicate that the upstream receptor for yes activation through Wnts is unknown so far.
Figure 3
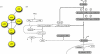
Modelling aspects of Wnt signalling by the use of ordinary differential equations. Part of the canonical Wnt signalling branch has been modelled in the Lee–Heinrich model (Lee et al. 2003). (a) Components of the network as indicated in figure 1_a_, which were included in the Lee–Heinrich model. In addition, dephosphorylation events, de novo synthesis and degradation of some components are included. (b) Reaction scheme included in the Lee–Heinrich model. Note that just a very small aspect of the Wnt signalling network has been implemented in the Lee–Heinrich model.
Figure 4
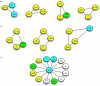
Network motifs of direct protein–protein interactions found within the Wnt signalling network. Examples for (a) triads and (b) tetrads of interactions identified. (c) Major components of the network, i.e. dishevelled represent hubs. Not all known interactions of dsh are shown. Additional interaction partners include Frodo/Dapper, Iday, Naked, MuSK, Eps8, Par-1, PAK and PP2C. Other hubs are LEF/TCF, β-catenin, axin or Frizzled.
Similar articles
- Wnt and calcium signaling: beta-catenin-independent pathways.
Kohn AD, Moon RT. Kohn AD, et al. Cell Calcium. 2005 Sep-Oct;38(3-4):439-46. doi: 10.1016/j.ceca.2005.06.022. Cell Calcium. 2005. PMID: 16099039 Review. - Network-based approaches for extending the Wnt signalling pathway and identifying context-specific sub-networks.
Saha S, Roman T, Galante A, Koyutürk M, Ewing RM. Saha S, et al. Int J Comput Biol Drug Des. 2012;5(3-4):185-205. doi: 10.1504/IJCBDD.2012.049203. Epub 2012 Sep 24. Int J Comput Biol Drug Des. 2012. PMID: 23013649 - Mathematical modelling of Wnt/β-catenin signalling.
Kofahl B, Wolf J. Kofahl B, et al. Biochem Soc Trans. 2010 Oct;38(5):1281-5. doi: 10.1042/BST0381281. Biochem Soc Trans. 2010. PMID: 20863299 - WNT signaling pathway and stem cell signaling network.
Katoh M, Katoh M. Katoh M, et al. Clin Cancer Res. 2007 Jul 15;13(14):4042-5. doi: 10.1158/1078-0432.CCR-06-2316. Clin Cancer Res. 2007. PMID: 17634527 Review.
Cited by
- Wnt Pathway in Bone Repair and Regeneration - What Do We Know So Far.
Houschyar KS, Tapking C, Borrelli MR, Popp D, Duscher D, Maan ZN, Chelliah MP, Li J, Harati K, Wallner C, Rein S, Pförringer D, Reumuth G, Grieb G, Mouraret S, Dadras M, Wagner JM, Cha JY, Siemers F, Lehnhardt M, Behr B. Houschyar KS, et al. Front Cell Dev Biol. 2019 Jan 7;6:170. doi: 10.3389/fcell.2018.00170. eCollection 2018. Front Cell Dev Biol. 2019. PMID: 30666305 Free PMC article. Review. - Representing dynamic biological networks with multi-scale probabilistic models.
Groß A, Kracher B, Kraus JM, Kühlwein SD, Pfister AS, Wiese S, Luckert K, Pötz O, Joos T, Van Daele D, De Raedt L, Kühl M, Kestler HA. Groß A, et al. Commun Biol. 2019 Jan 17;2:21. doi: 10.1038/s42003-018-0268-3. eCollection 2019. Commun Biol. 2019. PMID: 30675519 Free PMC article. - A model of the onset of the senescence associated secretory phenotype after DNA damage induced senescence.
Meyer P, Maity P, Burkovski A, Schwab J, Müssel C, Singh K, Ferreira FF, Krug L, Maier HJ, Wlaschek M, Wirth T, Kestler HA, Scharffetter-Kochanek K. Meyer P, et al. PLoS Comput Biol. 2017 Dec 4;13(12):e1005741. doi: 10.1371/journal.pcbi.1005741. eCollection 2017 Dec. PLoS Comput Biol. 2017. PMID: 29206223 Free PMC article. - Epigenetic regulation of WNT signaling in chronic lymphocytic leukemia.
Bennett LB, Taylor KH, Arthur GL, Rahmatpanah FB, Hooshmand SI, Caldwell CW. Bennett LB, et al. Epigenomics. 2010 Feb 1;2(1):53-70. doi: 10.2217/epi.09.43. Epigenomics. 2010. PMID: 20473358 Free PMC article. - Spatio-temporal regulation of Wnt and retinoic acid signaling by tbx16/spadetail during zebrafish mesoderm differentiation.
Mueller RL, Huang C, Ho RK. Mueller RL, et al. BMC Genomics. 2010 Sep 9;11:492. doi: 10.1186/1471-2164-11-492. BMC Genomics. 2010. PMID: 20828405 Free PMC article.
References
- Ahumada A, Slusarski D.C, Liu X, Moon R.T, Malbon C.C, Wang H.Y. Signaling of rat Frizzled-2 through phosphodiesterase and cyclic GMP. Science. 2002;298:2006–2010. doi:10.1126/science.1073776 - DOI - PubMed
- Albert I, Albert R. Conserved network motifs allow protein–protein interaction prediction. Bioinformatics. 2004;20:3346–3352. doi:10.1093/bioinformatics/bth402 - DOI - PubMed
- Albert R, Othmer H.G. The topology of the regulatory interactions predicts the expression pattern of the segment polarity genes in Drosophila melanogaster. J. Theor. Biol. 2003;223:1–18. doi:10.1016/S0022-5193(03)00035-3 - DOI - PMC - PubMed
- Alon U. Chapman & Hall/CRC; London, UK: 2007. An introduction to systems biology: design principles of biological circuits.
- Amonlirdviman K, Khare N.A, Tree D.R, Chen W.S, Axelrod J.D, Tomlin C.J. Mathematical modeling of planar cell polarity to understand domineering nonautonomy. Science. 2005;307:423–426. doi:10.1126/science.1105471 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous