An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses - PubMed (original) (raw)
Randomized Controlled Trial
. 2008 Jan 21;205(1):63-77.
doi: 10.1084/jem.20071331. Epub 2008 Jan 14.
Pierre-Alexandre Bart, Wolfgang Stöhr, Gonzalo Tapia, Miguel Garcia, Emmanuelle Medjitna-Rais, Séverine Burnet, Cristina Cellerai, Otto Erlwein, Tristan Barber, Christiane Moog, Peter Liljestrom, Ralf Wagner, Hans Wolf, Jean-Pierre Kraehenbuhl, Mariano Esteban, Jonathan Heeney, Marie-Joelle Frachette, James Tartaglia, Sheena McCormack, Abdel Babiker, Jonathan Weber, Giuseppe Pantaleo
Affiliations
- PMID: 18195071
- PMCID: PMC2234371
- DOI: 10.1084/jem.20071331
Randomized Controlled Trial
An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses
Alexandre Harari et al. J Exp Med. 2008.
Abstract
The EuroVacc 02 phase I trial has evaluated the safety and immunogenicity of a prime-boost regimen comprising recombinant DNA and the poxvirus vector NYVAC, both expressing a common immunogen consisting of Env, Gag, Pol, and Nef polypeptide domain from human immunodeficiency virus (HIV)-1 clade C isolate, CN54. 40 volunteers were randomized to receive DNA C or nothing on day 0 and at week 4, followed by NYVAC C at weeks 20 and 24. The primary immunogenicity endpoints were measured at weeks 26 and 28 by the quantification of T cell responses using the interferon gamma enzyme-linked immunospot assay. Our results indicate that the DNA C plus NYVAC C vaccine regimen was highly immunogenic, as indicated by the detection of T cell responses in 90% of vaccinees and was superior to responses induced by NYVAC C alone (33% of responders). The vaccine-induced T cell responses were (a) vigorous in the case of the env response (mean 480 spot-forming units/10(6) mononuclear cells at weeks 26/28), (b) polyfunctional for both CD4 and CD8 T cell responses, (c) broad (the average number of epitopes was 4.2 per responder), and (d) durable (T cell responses were present in 70% of vaccinees at week 72). The vaccine-induced T cell responses were strongest and most frequently directed against Env (91% of vaccines), but smaller responses against Gag-Pol-Nef were also observed in 48% of vaccinees. These results support the development of the poxvirus platform in the HIV vaccine field and the further clinical development of the DNA C plus NYVAC C vaccine regimen.
Figures
Figure 1.
RNA- and codon-optimized GPN and Env gene vector inserts. (A and B) Schematic representation of EV02 study design. ΔMyr, myristoylation-deficient; FS (-1), placing Gag and PolNef in one reading frame by removing the natural frameshift; ΔPR, protease-inactivated; RT-N, RT-C NH2 terminal and C-terminal part of the HIV reverse transcription; SC-Nef, scrambled Nef; RT-AS, active site of RT; SP, signal peptide.
Figure 2.
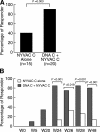
Immunogenicity of DNA C plus NYVAC C versus NYVAC C–alone vaccine regimens. (A) Percentage of responders in the two study groups at weeks 26/28, i.e., primary endpoints of the study. The percentage of responders was calculated on the basis of volunteers with a positive IFN-γ ELISPOT assay at weeks 26/28. (B) Percentage of responders at the different time points across the duration of the study.
Figure 3.
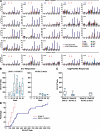
Magnitude of the vaccine-induced T cell responses. (A) Individual patterns of the T cell responses as measured by the frequencies of IFN-γ–secreting cells against different peptide pools encompassing the Env, Gag, Pol, and Nef proteins in an ELISPOT assay are shown for all responders in both study groups. Each bar corresponds to the reactivity against a different peptide pool, and positive responses (against Gag, Pol, and Nef) are indicated by a star. The Env-specific responses correspond to the sum of the mean of the responses induced after stimulation with the two Env peptide pools. (B) Median magnitude of vaccine-induced T cell responses against Env at different time points across the duration of the study. (C) Median magnitude of Gag-, Pol-, and Nef-specific vaccine-induced T cell responses at weeks 26 and 28. (D) Cumulative distribution of the sum of SFU/106 cells of positive responses at week 26 or 28 by randomization group. For each participant, the week of maximum T cell response (sum SFU) was chosen. Each step corresponds to the maximal IFN-γ ELISPOT value for each participant.
Figure 4.
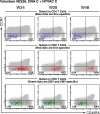
Vaccine-induced CD4 and CD8 T cell responses. (A) Flow cytometry profiles of vaccine-induced CD4 and CD8 T cell responses directed against Env in six out of the seven responders vaccinated with DNA C plus NYVAC C exhibiting CD4 and CD8 T cell responses. CD4 and CD8 T cell responses were defined using polychromatic flow cytometry. Blood mononuclear cells were stimulated with the relevant peptide pools and stained with IFN-γ, CD4, and CD8 antibodies. (B) Correlation of the IFN-γ–secreting cells measured by both flow cytometry and ELISPOT assay. This comparison was performed in the 17 volunteers with vaccine-induced T cell responses in the range of 100 SFU/106 cells or above.
Figure 5.
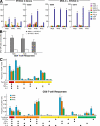
Functional profile of vaccine-induced CD4 and CD8 T cells. (A) Flow cytometry profiles of CD4 T cells and CD8 T cells able to mediate degranulation activity and to secrete IL-2, IFN-γ, and TNF-α are shown. Blood mononuclear cells were stimulated with the relevant peptide pools for 16 h and stained as described in Materials and methods. The functional profiles of both CD4 and CD8 T cells were performed on blood mononuclear cells of volunteers EU11, ES02, and ES08 that were vaccinated with DNA C plus NYVAC C. Volunteers ES02 and ES08 also exhibited Gag-specific in addition to Env-specific CD4 and CD8 T cell responses. With regard to the proliferation, cells were labeled with CFSE and stimulated with the relevant peptide pools, and proliferation was measured at day 6. (B) Functional composition of CD4 and CD8 T cell responses. The results shown were generated from the determinations in 11 responders. All the possible combinations of the responses are shown on the x axis, whereas the percentages of the functionally distinct cell populations within the total CD4 and CD8 T cell populations are shown on the y axis. Responses are grouped and color-coded on the basis of the number of functions. The pie chart summarizes the data, and each slice of the pie corresponds to the fraction of CD4 or CD8 T cells with a given number of functions within the total CD4 and CD8 T cell populations. Bars correspond to the fraction of different functionally distinct T cell populations within total CD4 and CD8 T cell populations. Mean and standard errors are also shown.
Figure 6.
Phenotypic analysis of vaccine-induced CD4 and CD8 T cells. Blood mononuclear cells obtained from volunteer ES26, that was vaccinated with DNA C plus NYVAC C, were stimulated with Env-derived peptide pools or with CMV/EBV-derived peptides for 16 h and stained with IL-2, IFN-γ, CD4, CD8, CD45RA, and CCR7 antibodies. The blue dots indicate Env-specific (IL-2 plus IFN-γ) vaccine-induced CD4 T cells. The red dots indicate Env-specific (IL-2 plus IFN-γ) vaccine-induced CD8 T cells. The green dots indicate CMV/EBV-specific (IL-2 plus IFN-γ) vaccine-induced CD8 T cells.
Figure 7.
Breadth of vaccine-induced T cell responses against Env. (A) Number of peptides/epitopes recognized in nine volunteers. Env-specific T cell responses were identified using peptide pools in preliminary experiments. The peptides of the relevant pool were then organized in a matrix setting to perform peptide/epitope mapping. Peptide/epitope mapping was performed combining the IFN-γ ELISPOT assay with polychromatic flow cytometry. (B) Example of a peptide/epitope mapping recognized by CD4 T cells. (C) Epitope mapping of the YSENSSEYY CD8 T cell epitope in two representative volunteers vaccinated with DNA C plus NYVAC C. (D) YSENSSEYY-specific CD8 T cells after staining with the relevant HLA-A*0101–YSENSSEYY pentameric complex.
Figure 8.
Durability of vaccine-induced T cell responses. (A) Monitoring of the T cell responses in representative volunteers of the two study groups. Only volunteers with positive T cell responses in the IFN-γ ELISPOT assay at week 48 were retested at week 72, which corresponded to 1 yr after the completion of the vaccination regimen. (B) Mean frequencies of IFN-γ–secreting cells/106 blood mononuclear cells at weeks 28, 48, and 72 in the eight volunteers within the DNA C plus NYVAC C group with a positive T cell response at weeks 72, 48, and 28, and mean frequencies of IFN-γ–secreting cells/106 blood mononuclear cells in the two volunteers within the NYVAC C group alone with responses at weeks 48 and 28. (C) Changes in the frequencies of functionally distinct CD4 and CD8 T cell populations over time. The results shown were generated from the determinations in six responders. The functional profile of vaccine-induced CD4 and CD8 T cells was assessed using polychromatic flow cytometry as described in Materials and methods. The functional composition of CD4 and CD8 T cell responses was determined as described in Fig. 5 B. All the possible combinations of the responses are shown on the x axis, whereas the percentage of the functionally distinct CD4 and CD8 T cell populations at weeks 28, 48, and 72 are shown on the y axis.
Comment in
- The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development?
Sekaly RP. Sekaly RP. J Exp Med. 2008 Jan 21;205(1):7-12. doi: 10.1084/jem.20072681. Epub 2008 Jan 14. J Exp Med. 2008. PMID: 18195078 Free PMC article.
Similar articles
- Virological and immunological characterization of novel NYVAC-based HIV/AIDS vaccine candidates expressing clade C trimeric soluble gp140(ZM96) and Gag(ZM96)-Pol-Nef(CN54) as virus-like particles.
Perdiguero B, Gómez CE, Cepeda V, Sánchez-Sampedro L, García-Arriaza J, Mejías-Pérez E, Jiménez V, Sánchez C, Sorzano CÓ, Oliveros JC, Delaloye J, Roger T, Calandra T, Asbach B, Wagner R, Kibler KV, Jacobs BL, Pantaleo G, Esteban M. Perdiguero B, et al. J Virol. 2015 Jan 15;89(2):970-88. doi: 10.1128/JVI.02469-14. Epub 2014 Oct 29. J Virol. 2015. PMID: 25355891 Free PMC article. - HIV/AIDS Vaccine Candidates Based on Replication-Competent Recombinant Poxvirus NYVAC-C-KC Expressing Trimeric gp140 and Gag-Derived Virus-Like Particles or Lacking the Viral Molecule B19 That Inhibits Type I Interferon Activate Relevant HIV-1-Specific B and T Cell Immune Functions in Nonhuman Primates.
García-Arriaza J, Perdiguero B, Heeney JL, Seaman MS, Montefiori DC, Yates NL, Tomaras GD, Ferrari G, Foulds KE, Roederer M, Self SG, Borate B, Gottardo R, Phogat S, Tartaglia J, Barnett SW, Burke B, Cristillo AD, Weiss DE, Lee C, Kibler KV, Jacobs BL, Wagner R, Ding S, Pantaleo G, Esteban M. García-Arriaza J, et al. J Virol. 2017 Apr 13;91(9):e02182-16. doi: 10.1128/JVI.02182-16. Print 2017 May 1. J Virol. 2017. PMID: 28179536 Free PMC article. - Potential To Streamline Heterologous DNA Prime and NYVAC/Protein Boost HIV Vaccine Regimens in Rhesus Macaques by Employing Improved Antigens.
Asbach B, Kliche A, Köstler J, Perdiguero B, Esteban M, Jacobs BL, Montefiori DC, LaBranche CC, Yates NL, Tomaras GD, Ferrari G, Foulds KE, Roederer M, Landucci G, Forthal DN, Seaman MS, Hawkins N, Self SG, Sato A, Gottardo R, Phogat S, Tartaglia J, Barnett SW, Burke B, Cristillo AD, Weiss DE, Francis J, Galmin L, Ding S, Heeney JL, Pantaleo G, Wagner R. Asbach B, et al. J Virol. 2016 Mar 28;90(8):4133-4149. doi: 10.1128/JVI.03135-15. Print 2016 Apr. J Virol. 2016. PMID: 26865719 Free PMC article. - Head-to-Head Comparison of Poxvirus NYVAC and ALVAC Vectors Expressing Identical HIV-1 Clade C Immunogens in Prime-Boost Combination with Env Protein in Nonhuman Primates.
García-Arriaza J, Perdiguero B, Heeney J, Seaman M, Montefiori DC, Labranche C, Yates NL, Shen X, Tomaras GD, Ferrari G, Foulds KE, McDermott A, Kao SF, Roederer M, Hawkins N, Self S, Yao J, Farrell P, Phogat S, Tartaglia J, Barnett SW, Burke B, Cristillo A, Weiss D, Lee C, Kibler K, Jacobs B, Asbach B, Wagner R, Ding S, Pantaleo G, Esteban M. García-Arriaza J, et al. J Virol. 2015 Aug;89(16):8525-39. doi: 10.1128/JVI.01265-15. Epub 2015 Jun 3. J Virol. 2015. PMID: 26041302 Free PMC article. - Poxvirus vectors as HIV/AIDS vaccines in humans.
Gómez CE, Perdiguero B, Garcia-Arriaza J, Esteban M. Gómez CE, et al. Hum Vaccin Immunother. 2012 Sep;8(9):1192-207. doi: 10.4161/hv.20778. Epub 2012 Aug 21. Hum Vaccin Immunother. 2012. PMID: 22906946 Free PMC article. Review.
Cited by
- Altering an artificial Gagpolnef polyprotein and mode of ENV co-administration affects the immunogenicity of a clade C HIV DNA vaccine.
Böckl K, Wild J, Bredl S, Kindsmüller K, Köstler J, Wagner R. Böckl K, et al. PLoS One. 2012;7(4):e34723. doi: 10.1371/journal.pone.0034723. Epub 2012 Apr 11. PLoS One. 2012. PMID: 22509350 Free PMC article. - Relevance of long-lived CD8(+) T effector memory cells for protective immunity elicited by heterologous prime-boost vaccination.
Vasconcelos JR, Dominguez MR, Araújo AF, Ersching J, Tararam CA, Bruna-Romero O, Rodrigues MM. Vasconcelos JR, et al. Front Immunol. 2012 Dec 4;3:358. doi: 10.3389/fimmu.2012.00358. eCollection 2012. Front Immunol. 2012. PMID: 23264773 Free PMC article. - Strategies for inducing effective neutralizing antibody responses against HIV-1.
Del Moral-Sánchez I, Sliepen K. Del Moral-Sánchez I, et al. Expert Rev Vaccines. 2019 Nov;18(11):1127-1143. doi: 10.1080/14760584.2019.1690458. Epub 2019 Dec 2. Expert Rev Vaccines. 2019. PMID: 31791150 Free PMC article. Review. - Strong HIV-specific CD4+ and CD8+ T-lymphocyte proliferative responses in healthy individuals immunized with an HIV-1 DNA vaccine and boosted with recombinant modified vaccinia virus ankara expressing HIV-1 genes.
Aboud S, Nilsson C, Karlén K, Marovich M, Wahren B, Sandström E, Gaines H, Biberfeld G, Godoy-Ramirez K. Aboud S, et al. Clin Vaccine Immunol. 2010 Jul;17(7):1124-31. doi: 10.1128/CVI.00008-10. Epub 2010 May 12. Clin Vaccine Immunol. 2010. PMID: 20463104 Free PMC article. Clinical Trial. - Highly persistent and effective prime/boost regimens against tuberculosis that use a multivalent modified vaccine virus Ankara-based tuberculosis vaccine with interleukin-15 as a molecular adjuvant.
Kolibab K, Yang A, Derrick SC, Waldmann TA, Perera LP, Morris SL. Kolibab K, et al. Clin Vaccine Immunol. 2010 May;17(5):793-801. doi: 10.1128/CVI.00006-10. Epub 2010 Mar 31. Clin Vaccine Immunol. 2010. PMID: 20357059 Free PMC article.
References
- Schwartländer, B., J. Stover, N. Walker, L. Bollinger, J.P. Gutierrez, W. McGreevey, M. Opuni, S. Forsythe, L. Kumaranayake, C. Watts, and S. Bertozzi. 2001. AIDS. Resource needs for HIV/AIDS. Science. 292:2434–2436. - PubMed
- Bart, P.A., A. Harari, and G. Pantaleo. 2006. Clinical studies of experimental vaccines. Current Opinion in HIV & AIDS. 1:286–293. - PubMed
- Letvin, N.L. 2006. Progress and obstacles in the development of an AIDS vaccine. Nat. Rev. Immunol. 6:930–939. - PubMed
- McMichael, A.J. 2006. HIV vaccines. Annu. Rev. Immunol. 24:227–255. - PubMed
- Heeney, J.L. 2004. Requirement of diverse T-helper responses elicited by HIV vaccines: induction of highly targeted humoral and CTL responses. Expert Rev. Vaccines. 3:S53–S64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials