Over-expression of CXCR4 on mesenchymal stem cells augments myoangiogenesis in the infarcted myocardium - PubMed (original) (raw)
Over-expression of CXCR4 on mesenchymal stem cells augments myoangiogenesis in the infarcted myocardium
Dongsheng Zhang et al. J Mol Cell Cardiol. 2008 Feb.
Abstract
Bone marrow mesenchymal stem cells (MSCs) participate in myocardial repair following myocardial infarction. However, their in vivo reparative capability is limited due to lack of their survival in the infarcted myocardium. To overcome this limitation, we genetically engineered male rat MSCs overexpressing CXCR4 in order to maximize the effect of stromal cell-derived factor-1alpha (SDF-1alpha) for cell migration and regeneration. MSCs were isolated from adult male rats and cultured. Adenoviral transduction was carried out to over-express either CXCR4/green fluorescent protein (Ad-CXCR4/GFP) or Ad-null/GFP alone (control). Flow cytometry was used to identify and isolate GFP/CXCR4 over-expressing MSCs for transplantation. Female rats were assigned to one of four groups (n=8 each) to receive GFP-transduced male MSCs (2 x 10(6)) via tail vein injection 3 days after ligation of the left anterior descending (LAD) coronary artery: GFP-transduced MSCs (Ad-null/GFP-MSCs, group 1) or MSCs over-expressing CXCR4/GFP (Ad-CXCR4/GFP-MSCs, group 2), or Ad-CXCR4/GFP-MSCs plus SDF-1alpha (50 ng/microl) (Ad-CXCR4/GFP-MSCs/SDF-1alpha, group 3), or Ad-miRNA targeting CXCR4 plus SDF-1alpha (Ad-miRNA/GFP-MSCs+SDF-1alpha treatment, group 4). Cardiodynamic data were obtained 4 weeks after induction of regional myocardial infarction (MI) using echocardiography after which hearts were harvested for immunohistochemical studies. The migration of GFP and Y-chromosome positive cells increased significantly in the peri- and infarct areas of groups 2 and 3 compared to control group (p<0.05), or miRNA-CXCR4 group (p<0.01). The number of CXCR4 positive cells in groups 2, 3 was intimately associated with angiogenesis and myogenesis. MSCs engraftment was blocked by pretreatment with miRNA (group 4). Cardiac function was significantly improved in rats receiving MSCs over-expressing CXCR4 alone or with SDF-1alpha. The up-regulation of matrix metalloproteinases (MMPs) by CXCR4 overexpressing MSCs perhaps facilitated their engraftment in the collagenous tissue of the infarcted area. CXCR4 over-expression led to enhance in vivo mobilization and engraftment of MSCs into ischemic area where these cells promoted neomyoangiogenesis and alleviated early signs of left ventricular remodeling.
Figures
Fig. 1
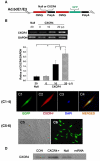
A: Generation of recombinant adenovirus vectors. B: RT-PCR for CXCR4 overexpression in MSCs. Panel B shows dose dependent CXCR4 overexpression. Null indicates Ad-null-GFP group. *p<0.05 vs. null group; †p<0.05 vs. CXCR4 (0 μl) group. Values mean±S.E.M., _n_=6 for each group. C(1–4): Immunofluorescence staining for EGFP and CXCR4. Panel C1 shows EGFP positive MSC, C2: Same MSC as “C1” except CXCR4 antibody staining (red color); C3: Same MSC as “C1” except nuclear staining by DAPI (blue color); C4, Same MSC as “C3” except DAPI co-localized with CXCR4 antibody staining and EGFP. Color codes for antibody staining are indicated. Panels C5–6 show the uniform transfection of MSCs with GFP and CXCR4 genes. C5, Non-transfected MSCs; C6, Adenovirus mediated EGFP and CXCR4 MSCs. Most of MSCs are uniformly transfected with EGFP and CXCR4 genes. D: CXCR4 expression on the membrane was determined by Western blotting using MSCs membrane extraction. CON indicates control group; CXCR4+, Ad-CXCR4/GFP group; Null, Ad-null-GFP group; miRNA, AdmiRNA targeting CXCR4 group. E–H: Flow cytometry of MSCs. Ad-null characterized cell fraction expressing CXCR4 (E) and c-kit (F); Ad-CXCR4 (G) and (H). Horizontal bar of each label in panel represents subpopulation of MSCs.
Fig. 1
A: Generation of recombinant adenovirus vectors. B: RT-PCR for CXCR4 overexpression in MSCs. Panel B shows dose dependent CXCR4 overexpression. Null indicates Ad-null-GFP group. *p<0.05 vs. null group; †p<0.05 vs. CXCR4 (0 μl) group. Values mean±S.E.M., _n_=6 for each group. C(1–4): Immunofluorescence staining for EGFP and CXCR4. Panel C1 shows EGFP positive MSC, C2: Same MSC as “C1” except CXCR4 antibody staining (red color); C3: Same MSC as “C1” except nuclear staining by DAPI (blue color); C4, Same MSC as “C3” except DAPI co-localized with CXCR4 antibody staining and EGFP. Color codes for antibody staining are indicated. Panels C5–6 show the uniform transfection of MSCs with GFP and CXCR4 genes. C5, Non-transfected MSCs; C6, Adenovirus mediated EGFP and CXCR4 MSCs. Most of MSCs are uniformly transfected with EGFP and CXCR4 genes. D: CXCR4 expression on the membrane was determined by Western blotting using MSCs membrane extraction. CON indicates control group; CXCR4+, Ad-CXCR4/GFP group; Null, Ad-null-GFP group; miRNA, AdmiRNA targeting CXCR4 group. E–H: Flow cytometry of MSCs. Ad-null characterized cell fraction expressing CXCR4 (E) and c-kit (F); Ad-CXCR4 (G) and (H). Horizontal bar of each label in panel represents subpopulation of MSCs.
Fig. 2
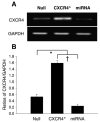
Effects of micro-RNA (miRNA) on MSCs. RT-PCR shows the transfection efficiency of CXCR4-miRNA on MSCs (A). CXCR4 expression was knocked down by miRNA targeting CXCR4 gene (B). Values are mean±S.E.M., n_=6 for each group. *p<0.05 vs. null group; †_p<0.01 vs. miRNA group.
Fig. 3
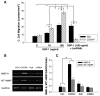
Cell migration and MMPs expression. A: Chemotaxis experiments were carried out by using transwells (5-μm pore). Cultured MSCs transduced with AdCXCR4/GFP (1 × 105) or Ad-miRNA targeting CXCR4 in 100 μl medium were added to the upper chamber, and 600 μl medium with or without SDF-1α (0, 10, 100 ng/ml) was placed in the bottom chamber. After 4 h incubation at 37 °C, migrating (bottom chamber) cells were counted. Exposure of Ad-CXCR4-MSCs to SDF-1α for 4 h resulted in strong cell migration, in a concentration dependent manner. The cell migration was significantly higher in Ad-CXCR4 group in the presence of SDF-1α (10 and 100 ng/ml) than in Ad-null group, p<0.05). The treatment of miRNA targeting CXCR4 prevented cell migration in response to SDF-1α, suggesting that CXCR4+-MSCs was functional and responded to SDF-1α. All values were expressed as mean±S.E.M. n_=6 for each group. *p<0.05 vs. SDF-1α (0 ng/ml) group; †_p<0.01 vs. other groups; #p<0.05 vs. null group. B: RT-PCR shows MMP-9 and MT1-MMP mRNA expression in CXCR4 overexpressing MSCs. CON indicates control MSCs group (lane 1); CXCR4+, CXCR4 overexpressing MSC group (lane 2); Null, adenoviral null treated MSCs (lane 3); miRNA, miRNA targeting CXCR4 (lane 4). GAPDH was used as the mRNA internal control to ensure equivalent loading. C: Quantitative assessment of CXCR4+-MSCs mRNA expression. All values were expressed as mean±S.E.M. _n_=6 for each group. *p<0.05 vs. control group.
Fig. 4
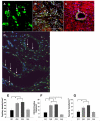
Migration and differentiation of MSCs in the infarcted myocardium. Panels A–D shows immunofluorescent staining in group 3 (data in other groups not shown). A: Immunostaining for EGFP-positive MSCs which migrated to the border zone of infarcted myocardium (green fluorescence, arrow). B: Same section as “A” except all nuclei (arrow) were identified by DAPI; C: Same section as “A and B” except α-sarcomeric actin staining for cardiac myocytes (red fluorescence); D: Same section as “A, B and C” except for colocalization of α-sarcomeric actin, EGFP and DAPI. Magnification × 200. Panels E–H shows FISH analysis for Y-chromosome. The presence of Y-chromosome positive nuclei (arrow) in the differentiated cardiomyocytes in group 1 (E), group 2 (F), group 3 (G) and group 4 (H). Myocytes were identified by sarcomeric α-actin (green). All nuclei were identified by DAPI staining (blue) (original magnification × 630, confocal imaging). I shows average number of transplanted GFP positive cells per unit area (mm2) in the peri-infarcted area after various treatments. J shows average number of transplanted Y-chromosome positive cells per unit area (mm2) in the peri-infarcted area after various treatments. All values were expressed as mean±S.E.M. n_=6 for each group. *p<0.05 vs. control group; †_p<0.05 vs. other groups. G1 indicates group 1; G2, group 2; G3, group 3; G4, group 4.
Fig. 5
Assessment of blood vessel density. Blood vessels were stained with CD31 (A), α-smooth muscle actin (SMA) (B) and Y-chromosome positive nuclei (D). Capillary density was identified by CD31 antibody in group 3 (A, arrows) and quantified (E). Immunocytochemistry of the heart tissues with polyclonal antibody revealed an intense staining of CXCR4 predominantly in the endothelial cells in the damaged myocardium in group 2 (C, arrow). D shows the presence of Ychromosome positive nuclei (arrow) in the differentiated blood vessels in group 3 (data in other groups not shown). DAPI-stained nuclei (blue) (original magnification × 630, confocal imaging). F shows CXCR4 positive vessels per unit area (mm2) in the infarcted area of various treatment groups. The number of capillary vessels located in the infarcted area is shown in group 3 (E, green color). F shows quantitative analysis of vascular density (anti α-smooth muscle actin antibody staining) was determined by various treatments. All values were expressed as mean±S.E.M. *p<0.05 vs. control group; †p<0.05 vs. other groups. _n_=6 for each group. (Original magnification × 200 except confocal imaging.) G1 indicates group 1; G2, group 2; G3, group 3; G4, group 4.
Fig. 6
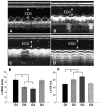
Assessment of cardiac function. M-mode echocardiograms are shown at 4 weeks in group 1 (A), group 2 (B), group 3 (C) and group 4 (D). Left ventricular enddiastolic diameters (LVDD, E) and ejection fraction (EF) (F) in hearts treated with CXCR4 overexpressing MSCs (G2), or SDF-1α plus CXCR4 overexpression (G3) was significantly better (F) than in control group (G1), or miRNA-CXCR4 group (G4). All values were expressed as mean±S.E.M. *p<0.05 vs. control group (G1). _n_=6 for each group.
Fig. 7
Measurement of fibrosis. Panel A shows sections for Masson-Trichome staining in various treatments. Hearts receiving CXCR4 overexpressing MSCs in group 2 (G2) or combination of CXCR4 overexpressing MSCs with SDF-1α in group 3 (G3) exhibited reduction in the fibrosis (arrow) as compared to group 1 (G1) or group 4 (G4) (A, G1–G4). Panel B shows percentage of fibrosis in hearts after permanent LAD occlusion and various treatments. Fibrosis in G2 and G3 was significantly reduced to 12.2±2.3%, 7.5±2.8%, respectively as compared with G1 (38.6±5.7%) or G4 (31.4±5.3%), B: All values were expressed as mean±S.E.M. *p<0.05 vs. control group; #p<0.05 vs. group 2 (_n_=6 in each group). G1 indicates group 1; G2, group 2; G3, group 3; G4, group 4.
Similar articles
- Genetically manipulated progenitor cell sheet with diprotin A improves myocardial function and repair of infarcted hearts.
Zhang D, Huang W, Dai B, Zhao T, Ashraf A, Millard RW, Ashraf M, Wang Y. Zhang D, et al. Am J Physiol Heart Circ Physiol. 2010 Nov;299(5):H1339-47. doi: 10.1152/ajpheart.00592.2010. Epub 2010 Aug 27. Am J Physiol Heart Circ Physiol. 2010. PMID: 20802132 Free PMC article. - Mesenchymal stem cells overexpressing CXCR4 attenuate remodeling of postmyocardial infarction by releasing matrix metalloproteinase-9.
Huang W, Wang T, Zhang D, Zhao T, Dai B, Ashraf A, Wang X, Xu M, Millard RW, Fan GC, Ashraf M, Yu XY, Wang Y. Huang W, et al. Stem Cells Dev. 2012 Mar 20;21(5):778-89. doi: 10.1089/scd.2011.0126. Epub 2011 Jul 22. Stem Cells Dev. 2012. PMID: 21671800 Free PMC article. - Stem cell homing and angiomyogenesis in transplanted hearts are enhanced by combined intramyocardial SDF-1alpha delivery and endogenous cytokine signaling.
Zhao T, Zhang D, Millard RW, Ashraf M, Wang Y. Zhao T, et al. Am J Physiol Heart Circ Physiol. 2009 Apr;296(4):H976-86. doi: 10.1152/ajpheart.01134.2008. Epub 2009 Jan 30. Am J Physiol Heart Circ Physiol. 2009. PMID: 19181961 Free PMC article. - SDF-1α as a therapeutic stem cell homing factor in myocardial infarction.
Ghadge SK, Mühlstedt S, Ozcelik C, Bader M. Ghadge SK, et al. Pharmacol Ther. 2011 Jan;129(1):97-108. doi: 10.1016/j.pharmthera.2010.09.011. Epub 2010 Oct 20. Pharmacol Ther. 2011. PMID: 20965212 Review. - Therapeutic strategies utilizing SDF-1α in ischaemic cardiomyopathy.
Ziff OJ, Bromage DI, Yellon DM, Davidson SM. Ziff OJ, et al. Cardiovasc Res. 2018 Mar 1;114(3):358-367. doi: 10.1093/cvr/cvx203. Cardiovasc Res. 2018. PMID: 29040423 Free PMC article. Review.
Cited by
- MicroRNAs in cancer treatment and prognosis.
Schoof CR, Botelho EL, Izzotti A, Vasques Ldos R. Schoof CR, et al. Am J Cancer Res. 2012;2(4):414-33. Epub 2012 Jun 28. Am J Cancer Res. 2012. PMID: 22860232 Free PMC article. - Challenges for heart disease stem cell therapy.
Hoover-Plow J, Gong Y. Hoover-Plow J, et al. Vasc Health Risk Manag. 2012;8:99-113. doi: 10.2147/VHRM.S25665. Epub 2012 Feb 17. Vasc Health Risk Manag. 2012. PMID: 22399855 Free PMC article. Review. - Stem cell recruitment after injury: lessons for regenerative medicine.
Rennert RC, Sorkin M, Garg RK, Gurtner GC. Rennert RC, et al. Regen Med. 2012 Nov;7(6):833-50. doi: 10.2217/rme.12.82. Regen Med. 2012. PMID: 23164083 Free PMC article. Review. - Mesenchymal stem/stromal cells as a delivery platform in cell and gene therapies.
D'souza N, Rossignoli F, Golinelli G, Grisendi G, Spano C, Candini O, Osturu S, Catani F, Paolucci P, Horwitz EM, Dominici M. D'souza N, et al. BMC Med. 2015 Aug 12;13:186. doi: 10.1186/s12916-015-0426-0. BMC Med. 2015. PMID: 26265166 Free PMC article. Review. - Combined therapy with atorvastatin and atorvastatin-pretreated mesenchymal stem cells enhances cardiac performance after acute myocardial infarction by activating SDF-1/CXCR4 axis.
Tian XQ, Yang YJ, Li Q, Xu J, Huang PS, Xiong YY, Li XD, Jin C, Qi K, Jiang LP, Chen GH, Qian L, Liu J, Geng YJ. Tian XQ, et al. Am J Transl Res. 2019 Jul 15;11(7):4214-4231. eCollection 2019. Am J Transl Res. 2019. PMID: 31396330 Free PMC article.
References
- Mangi AA, Noiseux N, Kong D, He H, Rezvani M, Ingwall JS, et al. Mesenchymal stem cells modified with Akt prevent remodeling and restore performance of infarcted hearts. Nat Med. 2003;9:1195–201. - PubMed
- Wojakowski W, Tendera M, Michalowska A, Majka M, Kucia M, Maslankiewicz K, et al. Mobilization of CD34/CXCR4+, CD34/CD117+, c-met+ stem cells, and mononuclear cells expressing early cardiac, muscle, and endothelial markers into peripheral blood in patients with acute myocardial infarction. Circulation. 2004;110(20):3213–20. - PubMed
- Kahn J, Byk T, Jansson-Sjostrand L, Petit I, Shivtiel S, Nagler A, et al. Overexpression of CXCR4 on human CD34+ progenitors increases their proliferation, migration, and NOD/SCID repopulation. Blood. 2004;103:2942–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL087861/HL/NHLBI NIH HHS/United States
- HL-081859-01/HL/NHLBI NIH HHS/United States
- HL083236/HL/NHLBI NIH HHS/United States
- HL-74272/HL/NHLBI NIH HHS/United States
- R01 HL081859/HL/NHLBI NIH HHS/United States
- HL-080686/HL/NHLBI NIH HHS/United States
- R01 HL080686/HL/NHLBI NIH HHS/United States
- R37 HL074272/HL/NHLBI NIH HHS/United States
- HL87861-01/HL/NHLBI NIH HHS/United States
- R01 HL083236/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical