Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes - PubMed (original) (raw)
Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes
David Moreira et al. BMC Evol Biol. 2008.
Abstract
Background: Although capable to evolve, viruses are generally considered non-living entities because they are acellular and devoid of metabolism. However, the recent publication of the genome sequence of the Mimivirus, a giant virus that parasitises amoebas, strengthened the idea that viruses should be included in the tree of life. In fact, the first phylogenetic analyses of a few Mimivirus genes that are also present in cellular lineages suggested that it could define an independent branch in the tree of life in addition to the three domains, Bacteria, Archaea and Eucarya.
Results: We tested this hypothesis by carrying out detailed phylogenetic analyses for all the conserved Mimivirus genes that have homologues in cellular organisms. We found no evidence supporting Mimivirus as a new branch in the tree of life. On the contrary, our phylogenetic trees strongly suggest that Mimivirus acquired most of these genes by horizontal gene transfer (HGT) either from its amoebal hosts or from bacteria that parasitise the same hosts. The detection of HGT events involving different eukaryotic donors suggests that the spectrum of hosts of Mimivirus may be larger than currently known.
Conclusion: The large number of genes acquired by Mimivirus from eukaryotic and bacterial sources suggests that HGT has been an important process in the evolution of its genome and the adaptation to parasitism.
Figures
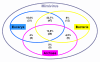
Figure 1
Taxonomic distribution of 128 conserved mimiviral ORFs. The number of homologues in the three domains of life (Eucarya, Archaea and Bacteria) is shown.
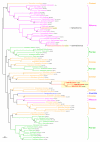
Figure 2
Bayesian phylogenetic trees of (A) the Zn-dependent alcohol dehydrogenase (MIMI_L498) and of (B) the putative outer membrane lipoprotein (MIMI_R877). These trees show HGT events from Proteobacteria that co-exist with Mimivirus within the same amoebal hosts. Numbers at nodes are Bayesian posterior probabilities. Scale bar represents the number of estimated changes per position for a unit of branch length.
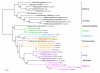
Figure 3
Bayesian phylogenetic tree of three serine/threonine protein kinases (MIMI_R818, MIMI_R826, MIMI_R831). The tree shows one gene acquisition by Mimivirus from its host, followed by two duplication events in the mimiviral lineage. Numbers at nodes are Bayesian posterior probabilities. Scale bar represents the number of estimated changes per position for a unit of branch length.
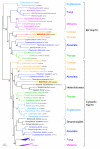
Figure 4
Bayesian phylogenetic tree of a cytosolic- and an endoplasmic reticulum (ER)-type HSP70 heat shock protein (MIMI_L254, and MIMI_L393). The tree shows the eukaryotic origin of the two mimiviral HSP70 by independent HGT from two distant eukaryotic groups (Amoebozoa and Heterolobosea). Numbers at nodes are Bayesian posterior probabilities. Scale bar represents the number of estimated changes per position for a unit of branch length.
Figure 5
Bayesian phylogenetic tree of the GDP mannose 4,6-dehydratase (MIMI_R141). The tree shows a case of gene acquisition by Mimivirus from a euglenozoan donor. Numbers at nodes are Bayesian posterior probabilities. Scale bar represents the number of estimated changes per position for a unit of branch length.
Similar articles
- Marine mimivirus relatives are probably large algal viruses.
Monier A, Larsen JB, Sandaa RA, Bratbak G, Claverie JM, Ogata H. Monier A, et al. Virol J. 2008 Jan 23;5:12. doi: 10.1186/1743-422X-5-12. Virol J. 2008. PMID: 18215256 Free PMC article. - Genomic and evolutionary aspects of Mimivirus.
Suzan-Monti M, La Scola B, Raoult D. Suzan-Monti M, et al. Virus Res. 2006 Apr;117(1):145-55. doi: 10.1016/j.virusres.2005.07.011. Epub 2005 Sep 21. Virus Res. 2006. PMID: 16181700 Review. - Study of Gene Trafficking between Acanthamoeba and Giant Viruses Suggests an Undiscovered Family of Amoeba-Infecting Viruses.
Maumus F, Blanc G. Maumus F, et al. Genome Biol Evol. 2016 Dec 14;8(11):3351-3363. doi: 10.1093/gbe/evw260. Genome Biol Evol. 2016. PMID: 27811174 Free PMC article. - Evolutionary genomics of nucleo-cytoplasmic large DNA viruses.
Iyer LM, Balaji S, Koonin EV, Aravind L. Iyer LM, et al. Virus Res. 2006 Apr;117(1):156-84. doi: 10.1016/j.virusres.2006.01.009. Epub 2006 Feb 21. Virus Res. 2006. PMID: 16494962 Review. - Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae.
Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. Arslan D, et al. Proc Natl Acad Sci U S A. 2011 Oct 18;108(42):17486-91. doi: 10.1073/pnas.1110889108. Epub 2011 Oct 10. Proc Natl Acad Sci U S A. 2011. PMID: 21987820 Free PMC article.
Cited by
- Giant viruses coexisted with the cellular ancestors and represent a distinct supergroup along with superkingdoms Archaea, Bacteria and Eukarya.
Nasir A, Kim KM, Caetano-Anolles G. Nasir A, et al. BMC Evol Biol. 2012 Aug 24;12:156. doi: 10.1186/1471-2148-12-156. BMC Evol Biol. 2012. PMID: 22920653 Free PMC article. - Comparative Genomics of Amphibian-like Ranaviruses, Nucleocytoplasmic Large DNA Viruses of Poikilotherms.
Price SJ. Price SJ. Evol Bioinform Online. 2016 Oct 25;11(Suppl 2):71-82. doi: 10.4137/EBO.S33490. eCollection 2015. Evol Bioinform Online. 2016. PMID: 27812275 Free PMC article. - Amoebae, Giant Viruses, and Virophages Make Up a Complex, Multilayered Threesome.
Diesend J, Kruse J, Hagedorn M, Hammann C. Diesend J, et al. Front Cell Infect Microbiol. 2018 Jan 11;7:527. doi: 10.3389/fcimb.2017.00527. eCollection 2017. Front Cell Infect Microbiol. 2018. PMID: 29376032 Free PMC article. Review. - 2b or Not 2b: Experimental Evolution of Functional Exogenous Sequences in a Plant RNA Virus.
Willemsen A, Zwart MP, Ambrós S, Carrasco JL, Elena SF. Willemsen A, et al. Genome Biol Evol. 2017 Feb 1;9(2):297-310. doi: 10.1093/gbe/evw300. Genome Biol Evol. 2017. PMID: 28137747 Free PMC article. - On the Evolution of the Mammalian Brain.
Torday JS, Miller WB Jr. Torday JS, et al. Front Syst Neurosci. 2016 Apr 19;10:31. doi: 10.3389/fnsys.2016.00031. eCollection 2016. Front Syst Neurosci. 2016. PMID: 27147985 Free PMC article.
References
- Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci USA. 2004;101:7716–7720. doi: 10.1073/pnas.0401773101. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources