Structural changes in TAF4b-TFIID correlate with promoter selectivity - PubMed (original) (raw)
Comparative Study
Structural changes in TAF4b-TFIID correlate with promoter selectivity
Wei-Li Liu et al. Mol Cell. 2008.
Abstract
Proper ovarian development requires the cell type-specific transcription factor TAF4b, a subunit of the core promoter recognition complex TFIID. We present the 35 A structure of a cell type-specific core promoter recognition complex containing TAF4b and TAF4 (4b/4-IID), which is responsible for directing transcriptional synergy between c-Jun and Sp1 at a TAF4b target promoter. As a first step toward correlating potential structure/function relationships of the prototypic TFIID versus 4b/4-IID, we have compared their 3D structures by electron microscopy and single-particle reconstruction. These studies reveal that TAF4b incorporation into TFIID induces an open conformation at the lobe involved in TFIIA and putative activator interactions. Importantly, this open conformation correlates with differential activator-dependent transcription and promoter recognition by 4b/4-IID. By combining functional and structural analysis, we find that distinct localized structural changes in a megadalton macromolecular assembly can significantly alter its activity and lead to a TAF4b-induced reprogramming of promoter specificity.
Figures
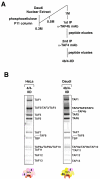
Figure 1. Isolation of cell type-specific 4b-IIDs
(A) Schematic representation of the purification procedure of 4b-IID from Daudi cells. 4b/4-IID was obtained by tandem immunoprecipitations from the fractions eluted from the phosphocellulose 1M KCl step using anti-TAF4b and anti-TAF4 antibodies. (B) Distinct TFIID complexes (i.e. HeLa 4/4-IID and Daudi 4b/4-IID) were analyzed by 4-12% SDS-PAGE (Invitrogen) and visualized by silver staining. The star indicates a weak band corresponding to TAF11.
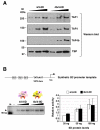
Figure 2. The basal transcriptional activities of distinct TFIID complexes
(A) The protein levels of 4/4- and 4b/4-IID complexes were examined by immunoblotting analysis using monoclonal antibodies against TAF1, TAF4, TAF4b and TBP. (B) Equivalent amounts (0, 20, 50, and 100 ng) of 4/4-IID and 4b/4-IID were added respectively into each reaction containing synthetic G3 promoter DNA template. The activities were quantified using a Typhoon scanner (Amersham Biosciences). Relative activities were calculated in comparison with the activity obtained from the lowest amount (20 ng) of 4/4-IID used. Each reaction was repeated several times and the representative data are shown (left panel). The arrows indicate the target transcripts. Error bars represent the standard deviations from at least three independent experiments (right panel).
Figure 3. Distinct transcriptional activities of TAF4b-target genes by 4b/4-IID
(A) Reactions were performed and quantitated exactly as described in Figure 2B, except that the native c-jun promoter template, a control hdm2 promoter (B), or native inhibin-βA subunit promoter (C) was utilized. The stars indicate non-specific transcripts. Error bars represent the standard deviations from at least three independent experiments.
Figure 4. Putative co-activator functions of 4b/4-IID
4b/4- or 4/4-IID (0 or 3 ng) were added to the transcription reactions containing c-jun, hdm2, or G3 promoter DNA templates in the absence or the presence of the activators c-Jun, Sp1, or both, respectively. The relative fold activation of individual TFIIDs were calculated based upon their relative activities that are measured in comparison to the activity with no addition of activators using the same promoter DNA. Error bars represent the standard deviations from at least three independent experiments.
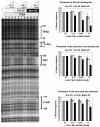
Figure 5. Elevated occupancy of activator binding sites in a 4b/4-IID dependent manner
Activator occupancy on the endogenous c-jun promoter was analyzed by DNase I footprinting assay. The binding elements for various transcription factors within the promoter region are indicated. Along with 20 nM of TFIIA, c-Jun (0, 12, 30 and 120 ng) and Sp1 (0, 8, 20, and 80 ng) were incubated respectively in the absence (lanes 1-4) or the presence of either 4/4-IID (lanes 5-8) or 4b/4-IID (lanes 9-12). Data shown are representative of at least three independent experiments. Quantification of the data is shown in the right panels. The percentage of relative DNAse I digestion was calculated based upon the digestions of individual binding elements in comparison to the digestions without addition of any activators. 0X, 0.1X, 0.25X, and 1X represent the increasing amounts of c-Jun and Sp1 protein levels as listed above. Error bars represent the standard deviations from at least three independent experiments.
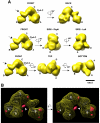
Figure 6. The 3D structure of 4b/4-IID and immunomapping of the TAF4b position within 4b/4-IID
(A) Different views of the 4b/4-IID EM structure with rotation angles are indicated. The major lobes are labeled as A, B, and C, and a smaller domain labeled as D lobe (see “BACK” view). The channel formed between the A and C lobes is labeled ChA-C. The arrow shown in the “FRONT” view indicates the undulating region located between the B and C lobes. The scale bar represents 100Å. (B) 3D difference density map between 4b/4-IID-anti-TAF4b and 4b/4-IID is shown in two different views. The yellow mesh corresponds to the 3D structure of 4b/4-IID with the A, B, C, and D lobes indicated. The extra densities (i.e. positive density differences) are shown in solid red. The most prominent difference region (indicated by the arrow) is located in the D lobe, close to C lobe and the central cavity, and was interpreted as the TAF4b antibody bound to 4b/4-IID.
Figure 7. Comparison of the 4b/4- and 4/4-IID 3D structures
Comparison from four different views (FRONT, BACK, SIDE, and TOP) of the 3D reconstructions of cell type-specific 4b/4-IID (left) and the canonical 4/4-IID (center). The distinct feature seen in the FRONT view is the ChA-C channel, which in 4/4-IID is significantly smaller than in 4b/4-IID (the curved arrows highlight this structural change). There is some missing density in 4b/4-IID located at the junction of A-C-D lobes (circles in the BACK view). Similarly, missing density was observed in 4/4-IID in the undulating region between B and C lobes (circles in the SIDE view). The right column shows the difference map between 4/4- and 4b/4-IID, with “red” representing positive differences and “green” representing negative differences. The “red” and “green” arrows point to the most significant positive and negative differences, respectively. The scale bar represents 100Å.
Similar articles
- Structure of promoter-bound TFIID and model of human pre-initiation complex assembly.
Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E. Louder RK, et al. Nature. 2016 Mar 31;531(7596):604-9. doi: 10.1038/nature17394. Epub 2016 Mar 23. Nature. 2016. PMID: 27007846 Free PMC article. - TAF4/4b x TAF12 displays a unique mode of DNA binding and is required for core promoter function of a subset of genes.
Gazit K, Moshonov S, Elfakess R, Sharon M, Mengus G, Davidson I, Dikstein R. Gazit K, et al. J Biol Chem. 2009 Sep 25;284(39):26286-96. doi: 10.1074/jbc.M109.011486. Epub 2009 Jul 27. J Biol Chem. 2009. PMID: 19635797 Free PMC article. - A transcription factor IIA-binding site differentially regulates RNA polymerase II-mediated transcription in a promoter context-dependent manner.
Wang J, Zhao S, He W, Wei Y, Zhang Y, Pegg H, Shore P, Roberts SGE, Deng W. Wang J, et al. J Biol Chem. 2017 Jul 14;292(28):11873-11885. doi: 10.1074/jbc.M116.770412. Epub 2017 May 24. J Biol Chem. 2017. PMID: 28539359 Free PMC article. - Promoter Recognition: Putting TFIID on the Spot.
Bhuiyan T, Timmers HTM. Bhuiyan T, et al. Trends Cell Biol. 2019 Sep;29(9):752-763. doi: 10.1016/j.tcb.2019.06.004. Epub 2019 Jul 9. Trends Cell Biol. 2019. PMID: 31300188 Review. - Transcription activation: unveiling the essential nature of TFIID.
Chen BS, Hampsey M. Chen BS, et al. Curr Biol. 2002 Sep 17;12(18):R620-2. doi: 10.1016/s0960-9822(02)01134-x. Curr Biol. 2002. PMID: 12372267 Review.
Cited by
- Molecular evolution of the testis TAFs of Drosophila.
Li VC, Davis JC, Lenkov K, Bolival B, Fuller MT, Petrov DA. Li VC, et al. Mol Biol Evol. 2009 May;26(5):1103-16. doi: 10.1093/molbev/msp030. Epub 2009 Feb 25. Mol Biol Evol. 2009. PMID: 19244474 Free PMC article. - Promoting developmental transcription.
Ohler U, Wassarman DA. Ohler U, et al. Development. 2010 Jan;137(1):15-26. doi: 10.1242/dev.035493. Development. 2010. PMID: 20023156 Free PMC article. Review. - Determining protein complex connectivity using a probabilistic deletion network derived from quantitative proteomics.
Sardiu ME, Gilmore JM, Carrozza MJ, Li B, Workman JL, Florens L, Washburn MP. Sardiu ME, et al. PLoS One. 2009 Oct 6;4(10):e7310. doi: 10.1371/journal.pone.0007310. PLoS One. 2009. PMID: 19806189 Free PMC article. - Targeting TBP-Associated Factors in Ovarian Cancer.
Ribeiro JR, Lovasco LA, Vanderhyden BC, Freiman RN. Ribeiro JR, et al. Front Oncol. 2014 Mar 11;4:45. doi: 10.3389/fonc.2014.00045. eCollection 2014. Front Oncol. 2014. PMID: 24653979 Free PMC article. Review. - p53 Dynamically Directs TFIID Assembly on Target Gene Promoters.
Coleman RA, Qiao Z, Singh SK, Peng CS, Cianfrocco M, Zhang Z, Piasecka A, Aldeborgh H, Basishvili G, Liu WL. Coleman RA, et al. Mol Cell Biol. 2017 Jun 15;37(13):e00085-17. doi: 10.1128/MCB.00085-17. Print 2017 Jul 1. Mol Cell Biol. 2017. PMID: 28416636 Free PMC article.
References
- Andel F, 3rd, Ladurner AG, Inouye C, Tjian R, Nogales E. Three-dimensional structure of the human TFIID-IIA-IIB complex. Science. 1999;286:2153–2156. - PubMed
- Chi T, Carey M. Assembly of the isomerized TFIIA--TFIID--TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. - PubMed
- Dikstein R, Zhou S, Tjian R. Human TAFII 105 is a cell type-specific TFIID subunit related to hTAFII130. Cell. 1996;87:137–146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 CA025417/CA/NCI NIH HHS/United States
- P01 CA112181/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- CA 25417/CA/NCI NIH HHS/United States
- R01 GM063072/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous