Argonaute loading improves the 5' precision of both MicroRNAs and their miRNA* strands in flies - PubMed (original) (raw)
Argonaute loading improves the 5' precision of both MicroRNAs and their miRNA* strands in flies
Hervé Seitz et al. Curr Biol. 2008.
Abstract
MicroRNAs (miRNAs) are short regulatory RNAs that direct repression of their mRNA targets. The miRNA "seed"-nucleotides 2-7-establishes target specificity by mediating target binding. Accurate processing of the miRNA 5' end is thought to be under strong selective pressure because a shift by just one nucleotide in the 5' end of a miRNA alters its seed sequence, redefining its repertoire of targets (Figure 1). Animal miRNAs are produced by the sequential cleavage of partially double-stranded precursors by the RNase III endonucleases Drosha and Dicer, thereby generating a transitory double-stranded intermediate comprising the miRNA paired to its partially complementary miRNA* strand. Here, we report that in flies, the 5' ends of miRNAs and miRNA* strands are typically more precisely defined than their 3' ends. Surprisingly, the precision of the 5' ends of both miRNA and miRNA* sequences increases after Argonaute2 (Ago2) loading. Our data imply that either many miRNA* sequences are under evolutionary pressure to maintain their seed sequences-that is, they have targets-or that secondary constraints, such as the sequence requirements for loading small RNAs into functional Argonaute complexes, narrow the range of miRNA and miRNA 5' ends that accumulate in flies.
Figures
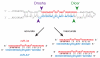
Figure 1. Inaccurate processing of the 5′ end of a miRNA alters its seed sequence
miRNA precursors are cleaved by two RNase III enzymes, Drosha and Dicer, liberating a short duplex: in this duplex, the mature miRNA (red) is paired to a partially complementary small RNA, the miRNA* (blue), derived from the opposite arm of the pre-miRNA stem. Inaccurate cleavage of the miRNA 5′ end changes its seed sequence (underlined).
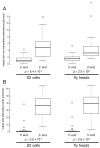
Figure 2. Cleavage inaccuracies are more frequent than non-templated additions
(A) The percentage of reads with non-templated 5′ or 3′ extensions was evaluated for each miRNA whose sequence was read at least 100 times. (B) The most abundant 5′ and 3′ ends were identified for each miRNA and all other ends corresponding to the sequence of the primary miRNA transcript were flagged as “alternative”. The percentage of reads with alternative ends was then determined for each miRNA read at least 100 times. Note the difference in the y-axis scales in (A) and (B). Box plots follow Tukey's standard conventions: a rectangle encloses all data from the first to the third quartiles, a bold horizontal line reports the median, whiskers connected to the rectangle indicate the largest and smallest non-outlier data, and outliers (values distant from the box by more than 1.5 times the interquartile range) are displayed as open circles.
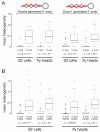
Figure 3. miRNA and miRNA* 5′ ends are more precisely defined than their 3′ ends
(A) miRNAs originating from the 5′ (left panels) or 3′ (right panels) arms of their pre-miRNAs were analyzed separately. For each miRNA, the heterogeneity of its termini was calculated as the mean of the absolute values of the distance between the 5′ or 3′ extremity of an individual templated read and the most abundant 5′ or 3′ ends for that miRNA. Sequences read from RNA isolated from fly heads and cultured S2 cells were analyzed separately. (B) Box-plots show the distribution of mean heterogeneity for the 5′ and 3′ ends of miRNA and miRNA* sequences.
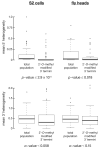
Figure 4. Ago2-loading, as evidenced by 3′ terminal 2′-_O_-methylation, refines miRNA and miRNA* 5′ ends
On average, the 5′ ends of the miRNAs and miRNA* strands in the 2′-_O_-methylated populations from both fly heads and S2 cells were more precisely defined than in the total population. We observed no statistically significant increase in the precision of the 3′ ends of the 3′ modified miRNAs and miRNA* strands.
Similar articles
- The 3'-to-5' exoribonuclease Nibbler shapes the 3' ends of microRNAs bound to Drosophila Argonaute1.
Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. Han BW, et al. Curr Biol. 2011 Nov 22;21(22):1878-87. doi: 10.1016/j.cub.2011.09.034. Epub 2011 Nov 3. Curr Biol. 2011. PMID: 22055293 Free PMC article. - Sorting of Drosophila small silencing RNAs.
Tomari Y, Du T, Zamore PD. Tomari Y, et al. Cell. 2007 Jul 27;130(2):299-308. doi: 10.1016/j.cell.2007.05.057. Cell. 2007. PMID: 17662944 Free PMC article. - Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties.
Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Wee LM, et al. Cell. 2012 Nov 21;151(5):1055-67. doi: 10.1016/j.cell.2012.10.036. Cell. 2012. PMID: 23178124 Free PMC article. - Argonaute-mediated translational repression (and activation).
Iwasaki S, Tomari Y. Iwasaki S, et al. Fly (Austin). 2009 Jul-Sep;3(3):204-6. Epub 2009 Jul 14. Fly (Austin). 2009. PMID: 19556851 Review. - Molecular mechanisms of RNA interference.
Wilson RC, Doudna JA. Wilson RC, et al. Annu Rev Biophys. 2013;42:217-39. doi: 10.1146/annurev-biophys-083012-130404. Annu Rev Biophys. 2013. PMID: 23654304 Free PMC article. Review.
Cited by
- The Smaug RNA-Binding Protein Is Essential for microRNA Synthesis During the Drosophila Maternal-to-Zygotic Transition.
Luo H, Li X, Claycomb JM, Lipshitz HD. Luo H, et al. G3 (Bethesda). 2016 Nov 8;6(11):3541-3551. doi: 10.1534/g3.116.034199. G3 (Bethesda). 2016. PMID: 27591754 Free PMC article. - Many roads to maturity: microRNA biogenesis pathways and their regulation.
Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Winter J, et al. Nat Cell Biol. 2009 Mar;11(3):228-34. doi: 10.1038/ncb0309-228. Nat Cell Biol. 2009. PMID: 19255566 Review. - Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans.
Chung WJ, Agius P, Westholm JO, Chen M, Okamura K, Robine N, Leslie CS, Lai EC. Chung WJ, et al. Genome Res. 2011 Feb;21(2):286-300. doi: 10.1101/gr.113050.110. Epub 2010 Dec 22. Genome Res. 2011. PMID: 21177960 Free PMC article. - Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae.
Biryukova I, Ye T, Levashina E. Biryukova I, et al. BMC Genomics. 2014 Jul 4;15(1):557. doi: 10.1186/1471-2164-15-557. BMC Genomics. 2014. PMID: 24997592 Free PMC article. - The 3'-to-5' exoribonuclease Nibbler shapes the 3' ends of microRNAs bound to Drosophila Argonaute1.
Han BW, Hung JH, Weng Z, Zamore PD, Ameres SL. Han BW, et al. Curr Biol. 2011 Nov 22;21(22):1878-87. doi: 10.1016/j.cub.2011.09.034. Epub 2011 Nov 3. Curr Biol. 2011. PMID: 22055293 Free PMC article.
References
- Lai EC. MicroRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. - PubMed
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. - PubMed
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. - PubMed
- Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. - PubMed
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM65236/GM/NIGMS NIH HHS/United States
- R01 GM062862/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM065236/GM/NIGMS NIH HHS/United States
- GM62862/GM/NIGMS NIH HHS/United States
- R37 GM062862/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases