TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program - PubMed (original) (raw)
Comparative Study
doi: 10.1371/journal.pgen.0040010. Epub 2007 Dec 13.
Lucinda K Southworth, Kavita Y Sarin, Andrew S Venteicher, Wenxiu Ma, Woody Chang, Peggie Cheung, Sohee Jun, Maja K Artandi, Naman Shah, Stuart K Kim, Steven E Artandi
Affiliations
- PMID: 18208333
- PMCID: PMC2211538
- DOI: 10.1371/journal.pgen.0040010
Comparative Study
TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program
Jinkuk Choi et al. PLoS Genet. 2008 Jan.
Abstract
Telomerase serves a critical role in stem cell function and tissue homeostasis. This role depends on its ability to synthesize telomere repeats in a manner dependent on the reverse transcriptase (RT) function of its protein component telomerase RT (TERT), as well as on a novel pathway whose mechanism is poorly understood. Here, we use a TERT mutant lacking RT function (TERT(ci)) to study the mechanism of TERT action in mammalian skin, an ideal tissue for studying progenitor cell biology. We show that TERT(ci) retains the full activities of wild-type TERT in enhancing keratinocyte proliferation in skin and in activating resting hair follicle stem cells, which triggers initiation of a new hair follicle growth phase and promotes hair synthesis. To understand the nature of this RT-independent function for TERT, we studied the genome-wide transcriptional response to acute changes in TERT levels in mouse skin. We find that TERT facilitates activation of progenitor cells in the skin and hair follicle by triggering a rapid change in gene expression that significantly overlaps the program controlling natural hair follicle cycling in wild-type mice. Statistical comparisons to other microarray gene sets using pattern-matching algorithms revealed that the TERT transcriptional response strongly resembles those mediated by Myc and Wnt, two proteins intimately associated with stem cell function and cancer. These data show that TERT controls tissue progenitor cells via transcriptional regulation of a developmental program converging on the Myc and Wnt pathways.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
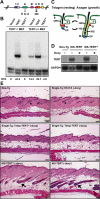
Figure 1. Conditional Expression of a Catalytically Inactive TERT Protein Induces the Anagen Phase of Hair Follicles
(A) Diagram of TERT protein structure shows domains conserved among all RTs. Three conserved aspartates (shown) are required for RT function. D702 in motif A was mutated to alanine, to create the mouse TERTci allele. (B) Mouse TERTci fails to reconstitute telomerase activity when transduced into TERT−/− MEFs and inhibits endogenous telomerase activity in TERT+/+ MEFs by TRAP. TRAP activities were quantified by measuring band intensities, and relative telomerase activity (RTA) is shown below each lane. (C) Hair follicles cycle between an active growth phase (anagen) and a resting phase (telogen). E, epidermis; ORS, outer root sheath; HS, hair shaft; M, matrix cells; DP, dermal papilla; IRS, inner root sheath; B, bulge stem cell niche; S, sebaceous gland. (D) Northern blot shows doxycycline-dependent suppression of TERT mRNA in dorsal skin from iK5-TERT and iK5-TERTci mice. (E) Induction of either TERTci or TERT mRNA in mouse epidermis initiates an anagen cycle between days 50–60. Doxycycline was withdrawn at day 10. Controls including non-transgenic, K5-tTA single transgenic, and tetop-TERTci and tetop-TERT single transgenic mice remained in telogen (asterisks). In contrast, iK5-TERTci and iK5-TERT mice are in anagen (black arrows). H&E, 100×.
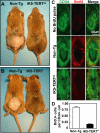
Figure 2. TERTci Promotes Hair Growth and Activates Hair Follicle Stem Cells
(A–B) Mice were shaved at approximately day 50 and hair growth was assessed after 2 wk. Note pink skin in shaved areas of controls, consistent with a continued telogen phase even after 14 d. (C) Dorsal skin biopsies were taken after long-term BrdU chase, and BrdU label retention in the CD34+ bulge stem cell compartment was assessed for non-Tg mice (middle) and iK5-TERTci mice (bottom). A non-Tg mouse that did not receive BrdU serves as a negative control (top). Green, CD34; red, BrdU; 400×. (D) Quantification of the number of BrdU positive cells per CD34+ cell in the bulge region. Conditional expression of TERTci depleted BrdU label from label retaining cells in the hair follicle bulge region. A total of 1,075 CD34+ cells from 75 bulges of four non-Tg mice, and 724 CD34+ cells from 65 bulges of six iK5-TERTci mice were analyzed. Error bars represent standard error.
Figure 3. Both TERT and TERTci Enhance Proliferation of Interfollicular Epidermis in Mouse Skin
(A) Expression of either TERT or TERTci increases thickness of interfollicular epidermis (IFE; black arrows). Dorsal skins were biopsied at days 55–60 and stained with H&E. Controls include non-transgenic and single-transgenic mice. (B) Quantification shows significant thickening of IFE in both iK5-TERT and iK5-TERTci (12 control, eight iK5-TERT, and 14 iK5-TERTci mice were analyzed. p < 0.00001 by one-way ANOVA and p < 0.05 by Student's _t_-test between control and iK5-TERT). (C,E) Expression of either TERT or TERTci increases proliferation in the basal layer of IFE. Abundance of Ki-67+ cells is increased in IFE of iK5-TERT and iK5-TERTci mice at days 55–60. Ki-67+ cells are stained by DAB (C) (black arrows), hematoxylin counterstain. Double immunostaining for Keratin 14 and Ki-67 (E). Green, K14; Red, Ki-67. (D,F) Quantification of Ki-67 staining shows significant increase in proliferation index. (D) Number of Ki-67+ cells per 100 μm. (F) Percentage of Ki-67+ cells within the basal layer of IFE (p < 0.0001 by one-way ANOVA and p < 0.01 by Student's _t_-test between control and iK5-TERT).
Figure 4. Acute Withdrawal of TERT Alters Expression of Genes Involved in Development, Signal Transduction, and Cell-to-Cell Signaling
(A) In two cohorts of iK5-TERT mice used for microarrays, TERT was switched on postnatally, inducing anagen at day 60. In TERT ON, or control samples, iK5-TERT mice remained off doxycycline, and TERT expression was maintained (green arrow). In TERT OFF samples, iK5-TERT mice were injected with doxycycline at t = 0, acutely silencing TERT expression (red arrow). (B) Northern blots show rapid silencing of TERT mRNA after doxycycline injection in iK5-TERT mice. GAPDH serves as a loading control. (C) Clustered heat map of TERT-regulated gene expression profile (FDR < 0.05). Control = TERT ON; +Doxy = TERT OFF (D–E) Venn diagrams depicting the distribution of TERT-activated genes (D) or TERT-repressed genes (E) within each functional category. (F) List of TERT-activated genes analyzed in (D). Red, components of Bmp or Wnt pathways; green, cell-cycle–related genes; blue, genes having mouse epidermal phenotypes. Note that not all genes with known functions are color-coded. (G) List of TERT-repressed genes analyzed in (E).
Figure 5. Validation of TERT-Regulated Genes by Quantitative RT-PCR
(A–B) Representative TERT-activated genes (A) and TERT-repressed genes (B) associated with different functional categories were validated by real-time PCR. (TERT ON = Control 24-h timepoint; TERT OFF = 24 h after doxycycline treatment timepoint; n = 3. Error bars represent standard errors). (C) Semi-quantitative RT-PCR validation of some representative genes. (D) Detailed results of quantitative RT-PCR validation of TERT-regulated genes.
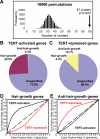
Figure 6. TERT Regulates a Concerted Transcriptional Program Overlapping Hair Growth and Anti–Hair Growth Genes in Normal Hair Cycling
(A) Histogram showing the frequency of genes whose start sites clustered using a randomly generated gene list equal in size to the TERT-regulated gene set (10,000 permutations). TERT-regulated genes reside in 87 clusters (black arrow), whereas only 33 clustered genes are expected. (B) TERT-activated genes are highly enriched with hair growth pattern genes (100 hair growth genes; 1 anti–hair growth gene; 287 others). (C) TERT-repressed genes are highly enriched with anti–hair growth genes (30 anti–hair growth genes; 1 hair growth gene; 206 others). (D–E) ROC plots of various TERT-activated gene lists and TERT-repressed gene lists, generated by different FDR thresholds, in predicting hair growth pattern genes (D) or anti–hair growth pattern genes (E).
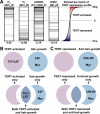
Figure 7. TERT-Regulated Genes Strongly Resemble Myc- and Wnt-Regulated Genes, and Are Enriched with TCF/LEF Binding Sites
(A) Representative GSEA results show that Myc and Wnt gene sets are highly enriched in our TERT-regulated dataset. Within each MSigDB gene set, horizontal black bars represent genes matching the rank ordered TERT-regulated dataset. Gene set names are denoted on top of each box and normalized enrichment scores (NES) on bottom. Note that negative NES values indicate enrichment in TERT-activated genes, and positive values indicate enrichment in TERT-repressed genes. (B–C) Graphic representation of _cis_-regulatory motif enrichment in various groups of genes. TCF/LEF sites were enriched in TERT-activated genes; E2F and Myc binding sites were enriched in hair growth genes (B); and CRE-BP and Myc sites were enriched in anti–hair growth genes (C). Although TCF/LEF sites were not over-represented in the entire collection of hair growth genes, they were significantly enriched in the subset of hair growth genes activated by TERT.
Similar articles
- Conditional telomerase induction causes proliferation of hair follicle stem cells.
Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Sarin KY, et al. Nature. 2005 Aug 18;436(7053):1048-52. doi: 10.1038/nature03836. Nature. 2005. PMID: 16107853 Free PMC article. - Clonal inactivation of TERT impairs stem cell competition.
Hasegawa K, Zhao Y, Garbuzov A, Corces MR, Neuhöfer P, Gillespie VM, Cheung P, Belk JA, Huang YH, Wei Y, Chen L, Chang HY, Artandi SE. Hasegawa K, et al. Nature. 2024 Aug;632(8023):201-208. doi: 10.1038/s41586-024-07700-w. Epub 2024 Jul 17. Nature. 2024. PMID: 39020172 Free PMC article. - Direct activation of TERT transcription by c-MYC.
Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Wu KJ, et al. Nat Genet. 1999 Feb;21(2):220-4. doi: 10.1038/6010. Nat Genet. 1999. PMID: 9988278 - Dissecting the non-canonical functions of telomerase.
Parkinson EK, Fitchett C, Cereser B. Parkinson EK, et al. Cytogenet Genome Res. 2008;122(3-4):273-80. doi: 10.1159/000167813. Epub 2009 Jan 30. Cytogenet Genome Res. 2008. PMID: 19188696 Review. - [Wnt signal transduction pathways and hair follicle stem cells].
Shao Y, Ni Z, Li Y. Shao Y, et al. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010 Aug;27(4):945-8. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 2010. PMID: 20842878 Review. Chinese.
Cited by
- Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo.
Hirashima K, Seimiya H. Hirashima K, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2022-32. doi: 10.1093/nar/gkv063. Epub 2015 Feb 4. Nucleic Acids Res. 2015. PMID: 25653161 Free PMC article. - PinX1: structure, regulation and its functions in cancer.
Li HL, Song J, Yong HM, Hou PF, Chen YS, Song WB, Bai J, Zheng JN. Li HL, et al. Oncotarget. 2016 Oct 4;7(40):66267-66275. doi: 10.18632/oncotarget.11411. Oncotarget. 2016. PMID: 27556185 Free PMC article. Review. - Telomerase deficiency delays renal recovery in mice after ischemia-reperfusion injury by impairing autophagy.
Cheng H, Fan X, Lawson WE, Paueksakon P, Harris RC. Cheng H, et al. Kidney Int. 2015 Jul;88(1):85-94. doi: 10.1038/ki.2015.69. Epub 2015 Mar 11. Kidney Int. 2015. PMID: 25760322 Free PMC article. - Means to the ends: The role of telomeres and telomere processing machinery in metastasis.
Robinson NJ, Schiemann WP. Robinson NJ, et al. Biochim Biophys Acta. 2016 Dec;1866(2):320-329. doi: 10.1016/j.bbcan.2016.10.005. Epub 2016 Oct 18. Biochim Biophys Acta. 2016. PMID: 27768860 Free PMC article. Review. - Telomerase deficiency affects normal brain functions in mice.
Lee J, Jo YS, Sung YH, Hwang IK, Kim H, Kim SY, Yi SS, Choi JS, Sun W, Seong JK, Lee HW. Lee J, et al. Neurochem Res. 2010 Feb;35(2):211-8. doi: 10.1007/s11064-009-0044-3. Epub 2009 Aug 15. Neurochem Res. 2010. PMID: 19685288
References
- Lee HW, Blasco MA, Gottlieb GJ, Horner JW, Jr, Greider CW, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. - PubMed
- Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–520. - PubMed
- Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. - PubMed
- Smogorzewska A, De Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007365/GM/NIGMS NIH HHS/United States
- P01 CA095616/CA/NCI NIH HHS/United States
- R01 CA111691/CA/NCI NIH HHS/United States
- GM07365/GM/NIGMS NIH HHS/United States
- R01 CA125453/CA/NCI NIH HHS/United States
- CA095616/CA/NCI NIH HHS/United States
- CA125453/CA/NCI NIH HHS/United States
- CA111691/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases