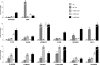The histone-like NF-Y is a bifunctional transcription factor - PubMed (original) (raw)
The histone-like NF-Y is a bifunctional transcription factor
Michele Ceribelli et al. Mol Cell Biol. 2008 Mar.
Abstract
NF-Y is a trimeric transcription factor containing H2A/H2B-like subunits, which specifically binds to the CCAAT box, a common eukaryotic promoter element. To gain insights into NF-Y-dependent transcriptional regulation, we assessed its relationships with positive histone marks by chromatin immunoprecipitation-on-chip and correlative-profiling studies. Unbiased identification of binding sites shows that the majority of genes are bound by NF-Y in the promoter and/or within the coding region. Parallel analysis of H3K9-14ac and H3K4me3 sites indicates that NF-Y loci can be divided in two distinct clusters: (i) a large cohort contains H3K9-14ac and H3K4me3 marks and correlates with expression and (ii) a sizeable group is devoid of these marks and is found on transcriptionally silent genes. Within this class, we find that NF-Y binding is associated with negative histone marks, such as H4K20me3 and H3K27me3. NF-Y removal by a dominant negative NF-YA leads to a decrease in the transcription of expressed genes associated with H3K4me3 and H3K9-14ac, while increasing the levels of many inactive genes. These data indicate that NF-Y is embedded in positive as well as in negative methyl histone marks, serving a dual function in transcriptional regulation, as an activator or as a repressor.
Figures
FIG. 1.
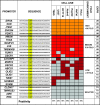
NF-Y binding to CCAAT promoters in vivo. Twenty-five randomly picked CCAAT promoters from chromosome 21 were analyzed by ChIP with anti-NF-YB antibody on chromatin derived from seven different cell lines. Enrichment over that for an irrelevant Flag control antibody was assessed by semiquantitative PCR and classified as high (>30-fold), medium (5- to 30-fold), or low (2- to 5-fold). Colored boxes indicate positivity and gray boxes indicate no enrichment in ChIPs. The bottom line refers to percentages of NF-Y-positive sites for each cell line. The expression patterns of the respective genes were analyzed according to available transcriptome data (UniGene build no. 186) and reported on the right. UB (ubiquitous) refers to genes expressed in at least 10 out of 45 body sites of UniGene's EST Profile Viewer, while TS (tissue specific) refers to genes showing expression in less than 10 body sites.
FIG. 2.
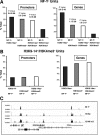
Categories and locations of NF-Y binding sites. (A) NF-Y promoter sites were plotted according to their positions with respect to the TSS. The frequencies of GE sites versus their distance from the TSS are also represented. EL sites were classified as present within 5 kb to the 5′ or 3′ end of an annotated gene, as present within a GenBank-annotated human mRNA not corresponding to a RefSeq (hmRNAs), or far from annotated genes (other). (B) Venn diagrams showing the overlap between NF-Y TUs containing a site in the promoter and those containing a site within the body of the gene.
FIG. 3.
CCAAT boxes in NF-Y locations. Average numbers of CCAAT boxes present in the different NF-Y location categories were plotted for the four stringencies initially considered. This number was calculated based on the actual mean dimensions of the identified NF-Y binding sites, ranging from 460 to 560 bp depending on the stringency, divided by the theoretical occurrence of the pentanucleotide CCAAT (once per 512 bp).
FIG. 4.
Correlation between NF-Y binding and active histone marks. (A) Percentages of NF-Y+ TUs that scored positive for either H3K9-K14ac or H3K4me3 or for both histone marks. (B) Percentages of H3K9-K14ac- or H3K4me3-positive TUs and of H3K9-K14ac/H3K4me3 double positives that shared at least one NF-Y location. (C) Position of NF-Y binding sites and histone modification islands within a representative cluster of five genes.
FIG. 5.
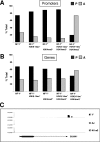
Correlation between NF-Y binding, active histone marks, and gene expression. (A) NF-Y+ promoters were scored for the presence (P) or absence (A) in the GSE6022 data set after the intersection with histone mark experiments. (B) Same as panel A, except that NF-Y+ genes were analyzed. (C) An NF-Y locus within the promoter of a representative nonexpressed gene, devoid of active histone marks, is shown.
FIG. 6.
ChIP with negative histone marks in nonexpressed NF-Y+ loci. Twelve NF-Y+/H3K9-K14ac−/H3K4me3− sites, randomly selected from the list of targets not expressed in HeLa cells (see File S7 in the supplemental material) were analyzed by ChIP with anti-NF-YB, anti-H3K27me3, anti-H3K9me3, and anti-H4K20me3 antibodies. Enrichment over that for an irrelevant Flag control antibody was assessed by semiquantitative PCR in duplicate experiments; enrichments greater than twofold were considered significant. The chromosome 11 satellite centromeric region and the promoter of the transcribed NF-Y target SON were used as internal controls.
FIG. 7.
Effects of NF-Y removal on gene expression. HeLa cells (left) were infected in parallel with control Ad-GFP, wild-type Ad-NF-YA, and the dominant negative Ad-YAm29 adenovirus. RT-PCR analyses of infected cells were performed in the linear range of amplification for the indicated genes. HCT116 cells (right) were analyzed under the same conditions. Western blot analysis confirmed equivalent overexpression of wild-type NF-YA and of YAm29 proteins (not shown).
Similar articles
- An NF-Y-dependent switch of positive and negative histone methyl marks on CCAAT promoters.
Donati G, Gatta R, Dolfini D, Fossati A, Ceribelli M, Mantovani R. Donati G, et al. PLoS One. 2008 Apr 30;3(4):e2066. doi: 10.1371/journal.pone.0002066. PLoS One. 2008. PMID: 18446193 Free PMC article. - NF-Y recruits Ash2L to impart H3K4 trimethylation on CCAAT promoters.
Fossati A, Dolfini D, Donati G, Mantovani R. Fossati A, et al. PLoS One. 2011 Mar 21;6(3):e17220. doi: 10.1371/journal.pone.0017220. PLoS One. 2011. PMID: 21445285 Free PMC article. - NF-Y affects histone acetylation and H2A.Z deposition in cell cycle promoters.
Gatta R, Mantovani R. Gatta R, et al. Epigenetics. 2011 Apr;6(4):526-34. doi: 10.4161/epi.6.4.14852. Epub 2011 Apr 1. Epigenetics. 2011. PMID: 21304275 - NF-Y and the transcriptional activation of CCAAT promoters.
Dolfini D, Gatta R, Mantovani R. Dolfini D, et al. Crit Rev Biochem Mol Biol. 2012 Jan-Feb;47(1):29-49. doi: 10.3109/10409238.2011.628970. Epub 2011 Nov 3. Crit Rev Biochem Mol Biol. 2012. PMID: 22050321 Review. - Structural determinants for NF-Y/DNA interaction at the CCAAT box.
Nardone V, Chaves-Sanjuan A, Nardini M. Nardone V, et al. Biochim Biophys Acta Gene Regul Mech. 2017 May;1860(5):571-580. doi: 10.1016/j.bbagrm.2016.09.006. Epub 2016 Sep 25. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27677949 Review.
Cited by
- NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis.
Li YJ, Fang Y, Fu YR, Huang JG, Wu CA, Zheng CC. Li YJ, et al. PLoS One. 2013 Apr 24;8(4):e61289. doi: 10.1371/journal.pone.0061289. Print 2013. PLoS One. 2013. PMID: 23637805 Free PMC article. - Novel function of U7 snRNA in the repression of HERV1/LTR12s and lincRNAs in human cells.
Plewka P, Szczesniak MW, Stepien A, Pasieka R, Wanowska E, Makalowska I, Raczynska KD. Plewka P, et al. Nucleic Acids Res. 2024 Sep 23;52(17):10504-10519. doi: 10.1093/nar/gkae738. Nucleic Acids Res. 2024. PMID: 39189459 Free PMC article. - NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors.
Fleming JD, Pavesi G, Benatti P, Imbriano C, Mantovani R, Struhl K. Fleming JD, et al. Genome Res. 2013 Aug;23(8):1195-209. doi: 10.1101/gr.148080.112. Epub 2013 Apr 17. Genome Res. 2013. PMID: 23595228 Free PMC article. - Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain.
Nowick K, Gernat T, Almaas E, Stubbs L. Nowick K, et al. Proc Natl Acad Sci U S A. 2009 Dec 29;106(52):22358-63. doi: 10.1073/pnas.0911376106. Epub 2009 Dec 10. Proc Natl Acad Sci U S A. 2009. PMID: 20007773 Free PMC article. - Genetic and functional assessment of the role of the rs13431652-A and rs573225-A alleles in the G6PC2 promoter that are strongly associated with elevated fasting glucose levels.
Bouatia-Naji N, Bonnefond A, Baerenwald DA, Marchand M, Bugliani M, Marchetti P, Pattou F, Printz RL, Flemming BP, Umunakwe OC, Conley NL, Vaxillaire M, Lantieri O, Balkau B, Marre M, Lévy-Marchal C, Elliott P, Jarvelin MR, Meyre D, Dina C, Oeser JK, Froguel P, O'Brien RM. Bouatia-Naji N, et al. Diabetes. 2010 Oct;59(10):2662-71. doi: 10.2337/db10-0389. Epub 2010 Jul 9. Diabetes. 2010. PMID: 20622168 Free PMC article.
References
- Azuara, V., P. Perry, S. Sauer, M. Spivakov, H. F. Jorgensen, R. M. John, M. Gouti, M. Casanova, G. Warnes, M. Merkenschlager, and A. G. Fisher. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8532-538. - PubMed
- Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. - PubMed
- Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources