A genome-wide screen for noncoding elements important in primate evolution - PubMed (original) (raw)
A genome-wide screen for noncoding elements important in primate evolution
Eliot C Bush et al. BMC Evol Biol. 2008.
Abstract
Background: A major goal in the study of human evolution is to identify key genetic changes which occurred over the course of primate evolution. According to one school of thought, many such changes are likely to be found in noncoding sequence. An approach to identifying these involves comparing multiple genomes to identify conserved regions with an accelerated substitution rate in a particular lineage. Such acceleration could be the result of positive selection.
Results: Here we develop a likelihood ratio test method to identify such regions. We apply it not only to the human terminal lineage, as has been done in previous studies, but also to a number of other branches in the primate tree. We present the top scoring elements, and compare our results with previous studies. We also present resequencing data from one particular element accelerated on the human lineage. These data indicate that the element lies in a region of low polymorphism in humans, consistent with the possibility of a recent selective sweep. They also show that the AT to GC bias for polymorphism in this region differs dramatically from that for substitutions.
Conclusion: Our results suggest that screens of this type will be helpful in unraveling the complex set of changes which occurred during primate evolution.
Figures
Figure 1
Illustration of methods. A. A phylogeny of the species we used with internal branches labeled. B. An element (green) and repeats (blue) within a 750 kb window. C. Constrained and unconstrained models. For each branch of the phylogeny, the element and the repeats are constrained to be proportional to each other. S is a tree wide scaling factor which accounts for the fact that elements vary in their general level of conservation (i.e. throughout the tree). The unconstrained model is the same as the constrained model, except that for whatever branch we are interested in (here the human terminal branch) we have added another variable F, which allows the element branch to be longer. We calculate the likelihood of the data under these two models. D. The constant of proportionality for each branch is based on the whole genome ratio between element and repeats for a particular branch.
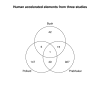
Figure 2
Overlap with previous studies. Venn diagram showing the overlap for human terminal branch elements from this study and two previous studies [6, 7, 39]. The group for the Pollard et al. study has and FDR adjusted p < 0.1, and the group for the Prabhakar study is p < 0.005.
Figure 3
An element on the human terminal branch. Multiple alignment and genomic context for human terminal branch element 2 (HAR2 in Pollard et al. 2006). In the alignment * indicates columns where human is derived.
Figure 4
An element on internal branch 2. Multiple alignment and genomic context for the fourth element on internal branch 2. In the alignment * indicates columns where catarhines are derived.
Similar articles
- Detecting positive selection within genomes: the problem of biased gene conversion.
Ratnakumar A, Mousset S, Glémin S, Berglund J, Galtier N, Duret L, Webster MT. Ratnakumar A, et al. Philos Trans R Soc Lond B Biol Sci. 2010 Aug 27;365(1552):2571-80. doi: 10.1098/rstb.2010.0007. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20643747 Free PMC article. - Human and non-human primate genomes share hotspots of positive selection.
Enard D, Depaulis F, Roest Crollius H. Enard D, et al. PLoS Genet. 2010 Feb 5;6(2):e1000840. doi: 10.1371/journal.pgen.1000840. PLoS Genet. 2010. PMID: 20140238 Free PMC article. - Detection of Pathways Affected by Positive Selection in Primate Lineages Ancestral to Humans.
Daub JT, Moretti S, Davydov II, Excoffier L, Robinson-Rechavi M. Daub JT, et al. Mol Biol Evol. 2017 Jun 1;34(6):1391-1402. doi: 10.1093/molbev/msx083. Mol Biol Evol. 2017. PMID: 28333345 Free PMC article. - Primate genomes.
Herlyn H, Zischler H. Herlyn H, et al. Genome Dyn. 2006;2:17-32. doi: 10.1159/000095090. Genome Dyn. 2006. PMID: 18753766 Review. - Fluorescence in situ hybridization to chromosomes as a tool to understand human and primate genome evolution.
Wienberg J. Wienberg J. Cytogenet Genome Res. 2005;108(1-3):139-60. doi: 10.1159/000080811. Cytogenet Genome Res. 2005. PMID: 15545725 Review.
Cited by
- Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior.
Doan RN, Bae BI, Cubelos B, Chang C, Hossain AA, Al-Saad S, Mukaddes NM, Oner O, Al-Saffar M, Balkhy S, Gascon GG; Homozygosity Mapping Consortium for Autism; Nieto M, Walsh CA. Doan RN, et al. Cell. 2016 Oct 6;167(2):341-354.e12. doi: 10.1016/j.cell.2016.08.071. Epub 2016 Sep 22. Cell. 2016. PMID: 27667684 Free PMC article. - Analysis of human accelerated DNA regions using archaic hominin genomes.
Burbano HA, Green RE, Maricic T, Lalueza-Fox C, de la Rasilla M, Rosas A, Kelso J, Pollard KS, Lachmann M, Pääbo S. Burbano HA, et al. PLoS One. 2012;7(3):e32877. doi: 10.1371/journal.pone.0032877. Epub 2012 Mar 7. PLoS One. 2012. PMID: 22412940 Free PMC article. - Selection on the regulation of sympathetic nervous activity in humans and chimpanzees.
Lee KS, Chatterjee P, Choi EY, Sung MK, Oh J, Won H, Park SM, Kim YJ, Yi SV, Choi JK. Lee KS, et al. PLoS Genet. 2018 Apr 19;14(4):e1007311. doi: 10.1371/journal.pgen.1007311. eCollection 2018 Apr. PLoS Genet. 2018. PMID: 29672586 Free PMC article. - Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex.
Boyd JL, Skove SL, Rouanet JP, Pilaz LJ, Bepler T, Gordân R, Wray GA, Silver DL. Boyd JL, et al. Curr Biol. 2015 Mar 16;25(6):772-779. doi: 10.1016/j.cub.2015.01.041. Epub 2015 Feb 19. Curr Biol. 2015. PMID: 25702574 Free PMC article. - Accelerated evolution of an Lhx2 enhancer shapes mammalian social hierarchies.
Wang Y, Dai G, Gu Z, Liu G, Tang K, Pan YH, Chen Y, Lin X, Wu N, Chen H, Feng S, Qiu S, Sun H, Li Q, Xu C, Mao Y, Zhang YE, Khaitovich P, Wang YL, Liu Q, Han JJ, Shao Z, Wei G, Xu C, Jing N, Li H. Wang Y, et al. Cell Res. 2020 May;30(5):408-420. doi: 10.1038/s41422-020-0308-7. Epub 2020 Apr 1. Cell Res. 2020. PMID: 32238901 Free PMC article.
References
- Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, Ibrahim M, Omar SA, Lema G, Nyambo TB, Ghori J, Bumpstead S, Pritchard JK, Wray GA, Deloukas P. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2006;39:31–40. doi: 10.1038/ng1946. - DOI - PMC - PubMed
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M, Vanderhaeghen P, Haussler D. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443(7108):167–172. doi: 10.1038/nature05113. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous