Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers - PubMed (original) (raw)
. 2008 Feb 5;105(5):1596-601.
doi: 10.1073/pnas.0711467105. Epub 2008 Jan 23.
Hélène Quach, Christine Harmant, Francesca Luca, Blandine Massonnet, Etienne Patin, Lucas Sica, Patrick Mouguiama-Daouda, David Comas, Shay Tzur, Oleg Balanovsky, Kenneth K Kidd, Judith R Kidd, Lolke van der Veen, Jean-Marie Hombert, Antoine Gessain, Paul Verdu, Alain Froment, Serge Bahuchet, Evelyne Heyer, Jean Dausset, Antonio Salas, Doron M Behar
Affiliations
- PMID: 18216239
- PMCID: PMC2234190
- DOI: 10.1073/pnas.0711467105
Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers
Lluís Quintana-Murci et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Two groups of populations with completely different lifestyles-the Pygmy hunter-gatherers and the Bantu-speaking farmers-coexist in Central Africa. We investigated the origins of these two groups and the interactions between them, by analyzing mtDNA variation in 1,404 individuals from 20 farming populations and 9 Pygmy populations from Central Africa, with the aim of shedding light on one of the most fascinating cultural transitions in human evolution (the transition from hunting and gathering to agriculture). Our data indicate that this region was colonized gradually, with an initial L1c-rich ancestral population ultimately giving rise to current-day farmers, who display various L1c clades, and to Pygmies, in whom L1c1a is the only surviving clade. Detailed phylogenetic analysis of complete mtDNA sequences for L1c1a showed this clade to be autochthonous to Central Africa, with its most recent branches shared between farmers and Pygmies. Coalescence analyses revealed that these two groups arose through a complex evolutionary process characterized by (i) initial divergence of the ancestors of contemporary Pygmies from an ancestral Central African population no more than approximately 70,000 years ago, (ii) a period of isolation between the two groups, accounting for their phenotypic differences, (iii) long-standing asymmetric maternal gene flow from Pygmies to the ancestors of the farming populations, beginning no more than approximately 40,000 years ago and persisting until a few thousand years ago, and (iv) enrichment of the maternal gene pool of the ancestors of the farming populations by the arrival and/or subsequent demographic expansion of L0a, L2, and L3 carriers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Phylogenetic tree of complete mtDNA sequences belonging to haplogroup L1c. The tree is rooted on Hg L1 and shows subhaplogroup affiliations. Mutations are shown on the branches. Transitions are labeled in uppercase letters, transversions are indicated in lowercase letters, deletions are indicated by a “d” after the deleted nucleotide position, and insertions are indicated by a dot, followed by the number and type of inserted nucleotides. Underlined nucleotide positions occur at least twice in the tree. The exclamation mark (!) at the end of a nucleotide position denotes a reversion to the ancestral state in the relative pathway from the rCRS (36). Individuals highlighted in orange correspond to PHG and those in blue to Bantu-speaking AGR. Population codes for each individual are as in Table 1. Coalescence age estimates for the main subhaplogroups are also reported.
Fig. 2.
PC plot based on haplogroup frequencies for the 29 population samples. For populations, codes, and geographic information, see Table 1.
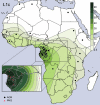
Fig. 3.
Spatial frequency distribution of haplogroup L1c in Africa. The interpolation map was constructed by using the frequencies of L1c, as observed in our dataset, and those retrieved from all published data on the African continent (
SI Tables 4 and 5
) excluding PHG populations (therefore corresponding to the distribution of L1c among AGR populations). The map of the frequency of L1c in PHG populations and the geographic location of these populations are presented in the projected portion of CA, as presented in the lower lefthand corner of the figure. The PHG interpolation map is shown separately because of the unique demographic nature of these populations.
Similar articles
- Insights into the demographic history of African Pygmies from complete mitochondrial genomes.
Batini C, Lopes J, Behar DM, Calafell F, Jorde LB, van der Veen L, Quintana-Murci L, Spedini G, Destro-Bisol G, Comas D. Batini C, et al. Mol Biol Evol. 2011 Feb;28(2):1099-110. doi: 10.1093/molbev/msq294. Epub 2010 Nov 1. Mol Biol Evol. 2011. PMID: 21041797 - Inferring the demographic history of African farmers and pygmy hunter-gatherers using a multilocus resequencing data set.
Patin E, Laval G, Barreiro LB, Salas A, Semino O, Santachiara-Benerecetti S, Kidd KK, Kidd JR, Van der Veen L, Hombert JM, Gessain A, Froment A, Bahuchet S, Heyer E, Quintana-Murci L. Patin E, et al. PLoS Genet. 2009 Apr;5(4):e1000448. doi: 10.1371/journal.pgen.1000448. Epub 2009 Apr 10. PLoS Genet. 2009. PMID: 19360089 Free PMC article. - Phylogeography of the human mitochondrial L1c haplogroup: genetic signatures of the prehistory of Central Africa.
Batini C, Coia V, Battaggia C, Rocha J, Pilkington MM, Spedini G, Comas D, Destro-Bisol G, Calafell F. Batini C, et al. Mol Phylogenet Evol. 2007 May;43(2):635-44. doi: 10.1016/j.ympev.2006.09.014. Epub 2006 Oct 5. Mol Phylogenet Evol. 2007. PMID: 17107816 - The demographic and adaptive history of central African hunter-gatherers and farmers.
Patin E, Quintana-Murci L. Patin E, et al. Curr Opin Genet Dev. 2018 Dec;53:90-97. doi: 10.1016/j.gde.2018.07.008. Epub 2018 Aug 10. Curr Opin Genet Dev. 2018. PMID: 30103089 Review. - African human mtDNA phylogeography at-a-glance.
Rosa A, Brehem A. Rosa A, et al. J Anthropol Sci. 2011;89:25-58. doi: 10.4436/jass.89006. Epub 2011 Mar 15. J Anthropol Sci. 2011. PMID: 21368343 Review.
Cited by
- Whole-genome sequencing reveals a complex African population demographic history and signatures of local adaptation.
Fan S, Spence JP, Feng Y, Hansen MEB, Terhorst J, Beltrame MH, Ranciaro A, Hirbo J, Beggs W, Thomas N, Nyambo T, Mpoloka SW, Mokone GG, Njamnshi A, Folkunang C, Meskel DW, Belay G, Song YS, Tishkoff SA. Fan S, et al. Cell. 2023 Mar 2;186(5):923-939.e14. doi: 10.1016/j.cell.2023.01.042. Cell. 2023. PMID: 36868214 Free PMC article. - African mitochondrial haplogroup L7: a 100,000-year-old maternal human lineage discovered through reassessment and new sequencing.
Maier PA, Runfeldt G, Estes RJ, Vilar MG. Maier PA, et al. Sci Rep. 2022 Jun 24;12(1):10747. doi: 10.1038/s41598-022-13856-0. Sci Rep. 2022. PMID: 35750688 Free PMC article. - Social ties in the Congo Basin: insights into tropical forest adaptation from BaYaka and their neighbours.
Boyette AH, Lew-Levy S, Jang H, Kandza V. Boyette AH, et al. Philos Trans R Soc Lond B Biol Sci. 2022 Apr 25;377(1849):20200490. doi: 10.1098/rstb.2020.0490. Epub 2022 Mar 7. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35249385 Free PMC article. Review. - Uniparental markers reveal new insights on subcontinental ancestry and sex-biased admixture in Brazil.
Joerin-Luque IA, Augusto DG, Calonga-Solís V, de Almeida RC, Lopes CVG, Petzl-Erler ML, Beltrame MH. Joerin-Luque IA, et al. Mol Genet Genomics. 2022 Mar;297(2):419-435. doi: 10.1007/s00438-022-01857-7. Epub 2022 Jan 21. Mol Genet Genomics. 2022. PMID: 35061071 - Genomic Landscape of the Mitochondrial Genome in the United Arab Emirates Native Population.
Aljasmi FA, Vijayan R, Sudalaimuthuasari N, Souid AK, Karuvantevida N, Almaskari R, Mohammed Abdul Kader H, Kundu B, Michel Hazzouri K, Amiri KMA. Aljasmi FA, et al. Genes (Basel). 2020 Aug 1;11(8):876. doi: 10.3390/genes11080876. Genes (Basel). 2020. PMID: 32752197 Free PMC article.
References
- Diamond J, Bellwood P. Farmers and their languages: The first expansions. Science. 2003;300:597–603. - PubMed
- Greenberg J. Linguistic evidence regarding Bantu origins. J African Hist. 1972;17:189–216.
- Newman J. The Peopling of Africa. New Haven, CT: Yale Univ Press; 1995.
- Phillipson D. African Archaeology. Cambridge, UK: Cambridge Univ Press; 1993.
- Cavalli-Sforza LL. In: African Pygmies. Cavalli-Sforza LL, editor. Orlando, FL: Academic; 1986. pp. 361–426.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources