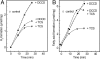The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features - PubMed (original) (raw)
. 2008 Feb 12;105(6):2128-33.
doi: 10.1073/pnas.0711093105. Epub 2008 Jan 24.
W Florian Fricke, Birgit Veith, Holger Brüggemann, Heiko Liesegang, Axel Strittmatter, Marcus Miethke, Wolfgang Buckel, Julia Hinderberger, Fuli Li, Christoph Hagemeier, Rudolf K Thauer, Gerhard Gottschalk
Affiliations
- PMID: 18218779
- PMCID: PMC2542871
- DOI: 10.1073/pnas.0711093105
The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features
Henning Seedorf et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Clostridium kluyveri is unique among the clostridia; it grows anaerobically on ethanol and acetate as sole energy sources. Fermentation products are butyrate, caproate, and H2. We report here the genome sequence of C. kluyveri, which revealed new insights into the metabolic capabilities of this well studied organism. A membrane-bound energy-converting NADH:ferredoxin oxidoreductase (RnfCDGEAB) and a cytoplasmic butyryl-CoA dehydrogenase complex (Bcd/EtfAB) coupling the reduction of crotonyl-CoA to butyryl-CoA with the reduction of ferredoxin represent a new energy-conserving module in anaerobes. The genes for NAD-dependent ethanol dehydrogenase and NAD(P)-dependent acetaldehyde dehydrogenase are located next to genes for microcompartment proteins, suggesting that the two enzymes, which are isolated together in a macromolecular complex, form a carboxysome-like structure. Unique for a strict anaerobe, C. kluyveri harbors three sets of genes predicted to encode for polyketide/nonribosomal peptide synthetase hybrides and one set for a nonribosomal peptide synthetase. The latter is predicted to catalyze the synthesis of a new siderophore, which is formed under iron-deficient growth conditions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Circular representation of the chromosome and plasmid of C. kluyveri. Genes encoded on the forward and reverse strands of the chromosome (outer two rings) are color-coded by red and dark blue, respectively. Predicted prophages are colored in green, and the sets of genes predicted to encode polyketide/nonribosomal peptide synthetases are indicated in yellow. The next inner ring shows the position of putative alien genes in gray. On the fourth ring the deviation from the average G+C content is plotted. Significant deviations are due to ribosomal RNA clusters (black arrows) and putative alien genes. Encoded transfer RNAs are indicated as black bars on the fifth ring. The plasmid map shows genes on the forward and reverse strands, color-coded in red and dark blue. The most inner ring shows the deviation from the average G+C content of the plasmid. Furthermore, the phage-terminase containing an intein is indicated in light blue. The intein was verified by heterologous production of the terminase in Escherichia coli (data not shown).
Fig. 2.
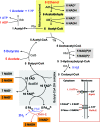
Ethanol-acetate fermentation of C. kluyveri. For simplification purposes it was assumed that only butyrate, acetic acid, and H2 rather than caproate are formed as fermentation products. Inset shows the two membrane-associated, energy-converting enzyme complexes involved in the fermentation: ferredoxin:NAD oxidoreductase (RnfA-E) and ATP synthase (AtpA-I). For the other cytoplasmic enzymes involved see
SI Table 2
. The clostridial ferredoxin with two [4Fe4S] clusters accepts two electrons upon reduction (28). In the scheme it is assumed that 7 of the 10 NADH required for the reduction of 5 crotonyl-CoA to 5 butyryl-CoA (see text and reaction 5) are generated during ethanol oxidation to acetyl-CoA (gray background) and that three are generated via NAD+ reduction with Fdred2− (orange background). It has been omitted that some of the Fdred2− generated during crotonyl-CoA reduction is also used for the regeneration of NADPH, which is required for acetoacetyl-CoA reduction. The yellow area indicates the microcompartment, and the shaded area highlights the steps proposed to be involved in the catalysis of reaction 5 (see text) by the butyryl-CoA dehydrogenase complex composed of butyryl-CoA dehydrogenase (Bcd) and the electron transport flavoproteins (EtfAB). Bcd, EtfA, and EtfB each contain FAD.
Fig. 3.
Effects of protonophore and F1Fo-ATPase inhibitor on H2 and fatty acid formation by cell suspensions of C. kluyveri. Shown is the effect of protonophore TCS and the F1Fo-ATPase inhibitor DCCD on hydrogen formation (A) and fatty acid formation (B). One mole of fatty acid corresponds to 1 mol of butyrate or 0.5 mol of caproate. For details on assay mixture composition see Materials and Methods. Open circles represent the control without TCS and DCCD (41).
Fig. 4.
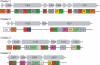
CDS clusters predicted to encode nonribosomal peptide synthetase and polyketide synthase-nonribosomal peptide synthetase hybrids. A, C, Cy, E, and T are domains of nonribosomal peptide synthetases: A, adenylation domain; C, condensation domain; Cy, cyclization domain; E, epimerization domain; T, thiolation domain. ACP, AT, KR, and KS are domains of polyketide synthases: ACP, acyl carrier protein domain; AT, acyltransferase domain; KR, ketoreductase domain; KS, ketosynthase domain. M, TE, and Sfp are domains occurring in both types: M, methyltransferase domain; TE, thioesterase domain; Sfp, phosphopantetheinyl transferase domain.
Similar articles
- NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron-bifurcating enzyme complex in Clostridium kluyveri.
Wang S, Huang H, Moll J, Thauer RK. Wang S, et al. J Bacteriol. 2010 Oct;192(19):5115-23. doi: 10.1128/JB.00612-10. Epub 2010 Jul 30. J Bacteriol. 2010. PMID: 20675474 Free PMC article. - Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri.
Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK. Li F, et al. J Bacteriol. 2008 Feb;190(3):843-50. doi: 10.1128/JB.01417-07. Epub 2007 Nov 9. J Bacteriol. 2008. PMID: 17993531 Free PMC article. - Rex in Clostridium kluyveri is a global redox-sensing transcriptional regulator.
Hu L, Huang H, Yuan H, Tao F, Xie H, Wang S. Hu L, et al. J Biotechnol. 2016 Sep 10;233:17-25. doi: 10.1016/j.jbiotec.2016.06.024. Epub 2016 Jun 29. J Biotechnol. 2016. PMID: 27373958 - Genome-scale metabolic reconstruction and analysis for Clostridium kluyveri.
Zou W, Ye G, Zhang J, Zhao C, Zhao X, Zhang K. Zou W, et al. Genome. 2018 Aug;61(8):605-613. doi: 10.1139/gen-2017-0177. Epub 2018 Jun 19. Genome. 2018. PMID: 29920212 - Recent advances in n-butanol and butyrate production using engineered Clostridium tyrobutyricum.
Bao T, Feng J, Jiang W, Fu H, Wang J, Yang ST. Bao T, et al. World J Microbiol Biotechnol. 2020 Aug 14;36(9):138. doi: 10.1007/s11274-020-02914-2. World J Microbiol Biotechnol. 2020. PMID: 32794091 Review.
Cited by
- Alternative sources of molybdenum for Methanococcus maripaludis and their implication for the evolution of molybdoenzymes.
Payne D, Keller LM, Larson J, Bothner B, Colman DR, Boyd ES. Payne D, et al. Commun Biol. 2024 Oct 16;7(1):1337. doi: 10.1038/s42003-024-07049-w. Commun Biol. 2024. PMID: 39414898 Free PMC article. - Urban sports fields support higher levels of soil butyrate and butyrate-producing bacteria than urban nature parks.
Brame JE, Liddicoat C, Abbott CA, Cando-Dumancela C, Fickling NW, Robinson JM, Breed MF. Brame JE, et al. Ecol Evol. 2024 Jul 22;14(7):e70057. doi: 10.1002/ece3.70057. eCollection 2024 Jul. Ecol Evol. 2024. PMID: 39041015 Free PMC article. - Variability in _n_-caprylate and _n_-caproate producing microbiomes in reactors with in-line product extraction.
Spirito CM, Lucas TN, Patz S, Jeon BS, Werner JJ, Trondsen LH, Guzman JJ, Huson DH, Angenent LT. Spirito CM, et al. mSystems. 2024 Aug 20;9(8):e0041624. doi: 10.1128/msystems.00416-24. Epub 2024 Jul 11. mSystems. 2024. PMID: 38990071 Free PMC article. - Simultaneous Formate and Syngas Conversion Boosts Growth and Product Formation by Clostridium ragsdalei.
Schwarz I, Angelina A, Hambrock P, Weuster-Botz D. Schwarz I, et al. Molecules. 2024 Jun 4;29(11):2661. doi: 10.3390/molecules29112661. Molecules. 2024. PMID: 38893534 Free PMC article. - The vast landscape of carbohydrate fermentation in prokaryotes.
Hackmann TJ. Hackmann TJ. FEMS Microbiol Rev. 2024 Jun 20;48(4):fuae016. doi: 10.1093/femsre/fuae016. FEMS Microbiol Rev. 2024. PMID: 38821505 Free PMC article. Review.
References
- Barker HA. The production of caproic and butyric acids by the methane fermentation of ethyl alcohol. Arch Mikrobiol. 1937;8:415–421.
- Thauer RK, Jungermann K, Wenning J, Decker K. Characterization of crotonate grown Clostridium kluyveri by its assimilatory metabolism. Arch Mikrobiol. 1968;64:125–129. - PubMed
- Thauer RK, Jungermann K, Henninger H, Wenning J, Decker K. The energy metabolism of Clostridium kluyveri. Eur J Biochem. 1968;4:173–180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases