MicroRNAs: biogenesis and molecular functions - PubMed (original) (raw)
Review
MicroRNAs: biogenesis and molecular functions
Xuhang Liu et al. Brain Pathol. 2008 Jan.
Abstract
Small regulatory RNAs are essential and ubiquitous riboregulators that are the key mediators of RNA interference (RNAi). They include microRNAs (miRNAs) and short-interfering RNAs (siRNAs), classes of approximately 22 nucleotide RNAs. miRNAs and siRNAs bind to Argonaute proteins and form effector complexes that regulate gene expression; in animals, this regulation occurs primarily at the post-transcriptional level. In this review, we will discuss our current understanding of how miRNA and siRNAs are generated and how they function to silence gene expression, focusing on animal and, in particular, mammalian miRNAs.
Figures
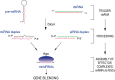
Figure 1
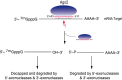
Biogenesis and function of microRNAs (miRNAs) and short‐interfering RNAs (siRNAs). As is true for almost all RNAs, miRNAs and siRNAs are derived from larger precursor RNAs. The precursor for miRNAs and siRNAs is double‐stranded (ds) RNA. In the case of miRNAs, the immediate precursor RNA is termed pre‐miRNA, adopts a hairpin structure and has a 5′‐phosphate and a 2‐nucleotide, 3′ overhang. In the case of siRNAs, the precursor is long dsRNA. Both are processed by the Dicer nuclease into duplexes termed miRNA or siRNA duplexes. The mature miRNA or siRNA is then released from the miRNA or siRNA duplex and binds to an Ago protein, to form a core effector complex that is commonly known as miRNP or RISC. miRNAs or siRNAs then act as specificity determinants to deposit the Ago proteins that they are bound to (RISCs, RNA‐induced silencing complex; miRNPs) onto their RNA targets, which are typically mRNAs. The deposited miRNP/RISC silences gene expression by suppressing the translation of the targeted mRNA and/or by destabilizing the targeted mRNA. Ago, Proteins that bind directly to miRNAs and siRNAs and are the core protein components of mitNPs/RISCs; Dicer, dsRNA endonuclease; miRNP, RiboNucleoProtein containing miRNA and an Ago protein.
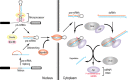
Figure 2
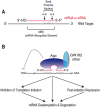
Overview of microRNA (miRNA) processing.
Figure 3
Precursor miRNA (Pre‐miRNA) processing: an RNA view.
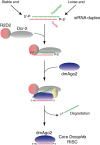
Figure 4
RNA‐induced silencing complex (RISC) assembly in Drosophila melanogaster.
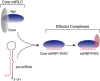
Figure 5
miRNP assembly in humans.
Figure 6
Target RNA cleavage by Ago2‐containing miRNP.
Figure 7
A. Principles of miRNA binding to target RNA. Binding of the miRNA to the target mRNA always occurs in the presence of an Ago protein; the protein was omitted to highlight the nucleotides that base‐pair. Base‐pairing between nucleotides 2 and 8 of an miRNA (an area known as “seed” or “proximal” or nucleus”) and its cognate mRNA target (MRE, miRNA recognition elements) is essential for binding of most miRNAs to their targets. Base‐pairing of other nucleotides of a miRNA and its target also occurs and may be required when complementarity in the proximal area is not perfect, or to enhance binding or function. B. Mechanisms of repression of targeted mRNA by miRNPs.
Similar articles
- Getting to the root of miRNA-mediated gene silencing.
Eulalio A, Huntzinger E, Izaurralde E. Eulalio A, et al. Cell. 2008 Jan 11;132(1):9-14. doi: 10.1016/j.cell.2007.12.024. Cell. 2008. PMID: 18191211 Review. - Diverse small non-coding RNAs in RNA interference pathways.
Li L, Liu Y. Li L, et al. Methods Mol Biol. 2011;764:169-82. doi: 10.1007/978-1-61779-188-8_11. Methods Mol Biol. 2011. PMID: 21748640 Review. - Post-transcriptional gene silencing by siRNAs and miRNAs.
Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Filipowicz W, et al. Curr Opin Struct Biol. 2005 Jun;15(3):331-41. doi: 10.1016/j.sbi.2005.05.006. Curr Opin Struct Biol. 2005. PMID: 15925505 Review. - Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs.
Wu L, Belasco JG. Wu L, et al. Mol Cell. 2008 Jan 18;29(1):1-7. doi: 10.1016/j.molcel.2007.12.010. Mol Cell. 2008. PMID: 18206964 Review. - Comparative analysis of argonaute-dependent small RNA pathways in Drosophila.
Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Zhou R, et al. Mol Cell. 2008 Nov 21;32(4):592-9. doi: 10.1016/j.molcel.2008.10.018. Mol Cell. 2008. PMID: 19026789 Free PMC article.
Cited by
- The genetics of hair cell development and regeneration.
Groves AK, Zhang KD, Fekete DM. Groves AK, et al. Annu Rev Neurosci. 2013 Jul 8;36:361-81. doi: 10.1146/annurev-neuro-062012-170309. Epub 2013 May 29. Annu Rev Neurosci. 2013. PMID: 23724999 Free PMC article. Review. - Prediction and characterization of microRNAs from eleven fish species by computational methods.
Huang Y, Zou Q, Ren HT, Sun XH. Huang Y, et al. Saudi J Biol Sci. 2015 Jul;22(4):374-81. doi: 10.1016/j.sjbs.2014.10.005. Epub 2014 Oct 23. Saudi J Biol Sci. 2015. PMID: 26150741 Free PMC article. - Gli3 is a novel downstream target of miR‑200a with an anti‑fibrotic role for progression of liver fibrosis in vivo and in vitro.
Li L, Ran J, Li L, Chen G, Zhang S, Wang Y. Li L, et al. Mol Med Rep. 2020 Apr;21(4):1861-1871. doi: 10.3892/mmr.2020.10997. Epub 2020 Feb 21. Mol Med Rep. 2020. PMID: 32319630 Free PMC article. - MicroRNAs as Molecular Targets for Cancer Therapy: On the Modulation of MicroRNA Expression.
Costa PM, Pedroso de Lima MC. Costa PM, et al. Pharmaceuticals (Basel). 2013 Sep 30;6(10):1195-220. doi: 10.3390/ph6101195. Pharmaceuticals (Basel). 2013. PMID: 24275848 Free PMC article. - Genome-wide exploration of miRNA function in mammalian muscle cell differentiation.
Polesskaya A, Degerny C, Pinna G, Maury Y, Kratassiouk G, Mouly V, Morozova N, Kropp J, Frandsen N, Harel-Bellan A. Polesskaya A, et al. PLoS One. 2013 Aug 21;8(8):e71927. doi: 10.1371/journal.pone.0071927. eCollection 2013. PLoS One. 2013. PMID: 23991007 Free PMC article.
References
- Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. - PubMed
- Ameres SL, Martinez J, Schroeder R (2007) Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130:101–112. - PubMed
- Aravin AA, Hannon GJ, Brennecke J (2007) The Piwi‐piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764. - PubMed
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R et al (2005) Regulation by let‐7 and lin‐4 miRNAs results in target mRNA degradation. Cell 122:553–563. - PubMed
- Bartel DP, Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5:396–400. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 NS053839/NS/NINDS NIH HHS/United States
- F30 NS054396/NS/NINDS NIH HHS/United States
- P30 HD026979/HD/NICHD NIH HHS/United States
- GM0720777/GM/NIGMS NIH HHS/United States
- P30-HD026979/HD/NICHD NIH HHS/United States
- F30NS054396/NS/NINDS NIH HHS/United States
- NS053839/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases