Design and use of fluorescent fusion proteins in cell biology - PubMed (original) (raw)
Design and use of fluorescent fusion proteins in cell biology
Erik Snapp. Curr Protoc Cell Biol. 2005 Jul.
Abstract
This unit describes strategies for designing functional fluorescent fusion protein constructs. Such constructs can be exploited as probes of cellular environments, protein dynamics, protein life histories, protein binding partners, and markers in living cells. The properties and uses of many currently available fluorescent proteins are discussed. In addition, alternative approaches and troubleshooting guidelines are provided.
Figures
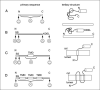
Figure 21.4.1
Appropriate positioning of a fluorescent protein (FP) in a fluorescent fusion protein construct. Preferred sites of FP fusion in the primary sequence of the protein of interest are indicated by happy face icons, and domains to be avoided are indicated by sad face icons. Each tertiary structure shows the folding of the sample construct with the FP (represented as a cylinder) fused at an optimal site. (A). A hypothetical globular protein expressed in the cytoplasm can have the FP fused at either the NH2− or the COOH-terminus. Typically, one end of the protein of interest will contain a functional domain that may be sterically hindered by an FP, and so it is useful to make both of the possible constructs. (B) A hypothetical lumenal protein contains an NH2−terminal signal sequence (SS), a mature domain, and a COOH-retention sequence (KDEL). An FP placed immediately after the SS or immediately before the retention sequence is less likely to interfere with the functioning of either sequence. (C) A single membrane–spanning protein has the additional constraint that the FP cannot be placed within or near the transmembrane domain (TMD), as this will disrupt the domain and cause problems with membrane integration. (D) A membrane multispanning protein has the same constraint as the example in panel C, but in multiple locations. The loops between the transmembrane domains are also poor choices, because the exact spacing between transmembrane domains is often important for protein folding, and because these loops often contain functional domains. Abbreviation: cyt, cytoplasm.
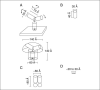
Figure 21.4.2
Relative sizes of (A) immunoglobulin G (IgG; reference for comparison with panels B to D), (B) green fluorescent protein (GFP), (C) the Discosoma red fluorescent protein (DsRed) tetramer, and (D) biarsenical tetracysteine.
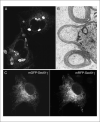
Figure 21.4.3
The fusion of a fluorescent protein (FP) to a native protein may change the protein's normal localization pattern or may lead to the formation of aggregates or oligomers. (A) The fusion of nonmonomerized enhanced green fluorescent protein (EGFP) to a resident endoplasmic reticulum (ER) membrane protein induces the formation of an organized smooth ER structure. (B) The fusion of monomerized EGFP to the same protein does not grossly alter the structure of the ER. (C) A Cos-7 cell expressing two fluorescent fusion proteins (FFPs), one containing monomerized green fluorescent protein (mGFP; left-hand image) and the other containing monomerized red fluorescent protein (mRFP; right-hand image). The mGFP-containing FFP localizes to the ER network, and similarly, the mRFP-containing FFP colocalizes to the ER membranes. The image yielded by the mRFP-tagged protein shows bright puncta, which are probably mRFP aggregates.
Similar articles
- Escherichia coli proteome chips for detecting protein-protein interactions.
Oishi Y, Yunomura S, Kawahashi Y, Doi N, Takashima H, Baba T, Mori H, Yanagawa H. Oishi Y, et al. Proteomics. 2006 Dec;6(24):6433-6. doi: 10.1002/pmic.200600341. Proteomics. 2006. PMID: 17109382 No abstract available. - Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis.
Hu CD, Grinberg AV, Kerppola TK. Hu CD, et al. Curr Protoc Cell Biol. 2006 Jan;Chapter 21:Unit 21.3. doi: 10.1002/0471143030.cb2103s29. Curr Protoc Cell Biol. 2006. PMID: 18228482 - Fluorescent labeling of recombinant proteins in living cells with FlAsH.
Griffin BA, Adams SR, Jones J, Tsien RY. Griffin BA, et al. Methods Enzymol. 2000;327:565-78. doi: 10.1016/s0076-6879(00)27302-3. Methods Enzymol. 2000. PMID: 11045009 No abstract available. - Methods for detection and measurement of hydrogen peroxide inside and outside of cells.
Rhee SG, Chang TS, Jeong W, Kang D. Rhee SG, et al. Mol Cells. 2010 Jun;29(6):539-49. doi: 10.1007/s10059-010-0082-3. Epub 2010 Jun 4. Mol Cells. 2010. PMID: 20526816 Review. - Out with the old, in with the new? Comparing methods for measuring protein degradation.
Yewdell JW, Lacsina JR, Rechsteiner MC, Nicchitta CV. Yewdell JW, et al. Cell Biol Int. 2011 May;35(5):457-62. doi: 10.1042/CBI20110055. Cell Biol Int. 2011. PMID: 21476986 Free PMC article. Review.
Cited by
- Imaging Subcellular Structures in the Living Zebrafish Embryo.
Engerer P, Plucinska G, Thong R, Trovò L, Paquet D, Godinho L. Engerer P, et al. J Vis Exp. 2016 Apr 2;(110):e53456. doi: 10.3791/53456. J Vis Exp. 2016. PMID: 27078038 Free PMC article. - Genetically Encoded Biosensors for the Fluorescence Detection of O2 and Reactive O2 Species.
Marchetti M, Ronda L, Cozzi M, Bettati S, Bruno S. Marchetti M, et al. Sensors (Basel). 2023 Oct 17;23(20):8517. doi: 10.3390/s23208517. Sensors (Basel). 2023. PMID: 37896609 Free PMC article. Review. - Production and cleavage of a fusion protein of porcine trypsinogen and enhanced green fluorescent protein (EGFP) in Pichia pastoris.
Raschmanová H, Paulová L, Branská B, Knejzlík Z, Melzoch K, Kovar K. Raschmanová H, et al. Folia Microbiol (Praha). 2018 Nov;63(6):773-787. doi: 10.1007/s12223-018-0619-y. Epub 2018 Jun 5. Folia Microbiol (Praha). 2018. PMID: 29872953 - Independent anterograde transport and retrograde cotransport of domain components of myelinated axons.
Bekku Y, Salzer JL. Bekku Y, et al. J Cell Biol. 2020 Jun 1;219(6):e201906071. doi: 10.1083/jcb.201906071. J Cell Biol. 2020. PMID: 32289157 Free PMC article. - Ectopic Expression of an Atypical Hydrophobic Group 5 LEA Protein from Wild Peanut, Arachis diogoi Confers Abiotic Stress Tolerance in Tobacco.
Sharma A, Kumar D, Kumar S, Rampuria S, Reddy AR, Kirti PB. Sharma A, et al. PLoS One. 2016 Mar 3;11(3):e0150609. doi: 10.1371/journal.pone.0150609. eCollection 2016. PLoS One. 2016. PMID: 26938884 Free PMC article.
References
- Albe KR, Butler MH, Wright BE. Cellular concentrations of enzymes and their substrates. J. Theor. Biol. 1990;143:163–195. - PubMed
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons; Hoboken, N.J.: 2005.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources