Systematic mutagenesis of the murine gammaherpesvirus 68 M2 protein identifies domains important for chronic infection - PubMed (original) (raw)
Systematic mutagenesis of the murine gammaherpesvirus 68 M2 protein identifies domains important for chronic infection
Jeremy H Herskowitz et al. J Virol. 2008 Apr.
Abstract
Murine gammaherpesvirus 68 (MHV68) infection of inbred mice represents a genetically tractable small-animal model for assessing the requirements for the establishment of latency, as well as reactivation from latency, within the lymphoid compartment. By day 16 postinfection, MHV68 latency in the spleen is found in B cells, dendritic cells, and macrophages. However, as with Epstein-Barr virus, by 3 months postinfection MHV68 latency is predominantly found in isotype-switched memory B cells. The MHV68 M2 gene product is a latency-associated antigen with no discernible homology to any known cellular or viral proteins. However, depending on experimental conditions, the M2 protein has been shown to play a critical role in both the efficient establishment of latency in splenic B cells and reactivation from latently infected splenic B cells. Inspection of the sequence of the M2 protein reveals several hallmarks of a signaling molecule, including multiple PXXP motifs and two potential tyrosine phosphorylation sites. Here, we report the generation of a panel of recombinant MHV68 viruses harboring mutations in the M2 gene that disrupt putative functional motifs. Subsequent analyses of the panel of M2 mutant viruses revealed a functionally important cluster of PXXP motifs in the C-terminal region of M2, which have previously been implicated in binding Vav proteins (P. A. Madureira, P. Matos, I. Soeiro, L. K. Dixon, J. P. Simas, and E. W. Lam, J. Biol. Chem. 280:37310-37318, 2005; L. Rodrigues, M. Pires de Miranda, M. J. Caloca, X. R. Bustelo, and J. P. Simas, J. Virol. 80:6123-6135, 2006). Further characterization of two adjacent PXXP motifs in the C terminus of the M2 protein revealed differences in the functions of these domains in M2-driven expansion of primary murine B cells in culture. Finally, we show that tyrosine residues 120 and 129 play a critical role in both the establishment of splenic latency and reactivation from latency upon explant of splenocytes into tissue culture. Taken together, these analyses will aide future studies for identifying M2 interacting partners and B-cell signaling pathways that are manipulated by the M2 protein.
Figures
FIG. 1.
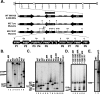
Deduced amino acid sequence of the M2 protein, with PXXP motifs and candidate tyrosine phosphorylation sites indicated in bold. Each PXXP motif is numbered, and the mutations introduced into each motif are indicated above the M2 sequence. The mutations of proline residues 70, 71, and 73 disrupt both the P3 and P4 PXXP motifs. Also shown are the mutations introduced into the tyrosine residues at positions 120 and 129, which were mutated to either phenylalanine or aspartic acid. The P7 PXXP motif is the only consensus class I SH3 binding motif (K/RxxPxxP) present in the M2 protein.
FIG. 2.
Construction of M2 mutant viruses and corresponding marker rescue viruses. (A) The MHV68 genome cloned as a BAC (2) was used to generate all M2 recombinant viruses by RecA-mediated recombination. For M2 mutant viruses exhibiting robust establishment-of-latency and/or reactivation-from-latency phenotypes, a marker rescue virus in which the wt M2 sequences were targeted to the M2 ORF by allelic exchange to restore the wt phenotype was generated. The DNA probe used for the Southern blot analyses contained sequence from the M2 ORF (genomic coordinates, bp 4031 to 4627). Relevant restriction sites are indicated. A, AluI; Bg, BglI; D, DdeI; N, NcoI; BU, BstUI; Bs, BssHII. (B and C) Southern blot analyses of recombinant viruses harboring mutations in the indicated PXXP motifs of M2, along with corresponding marker rescue viruses. (D) Southern blot analyses of recombinant viruses harboring mutations in the indicated tyrosine residues of M2 and their corresponding marker rescue viruses. (E) Southern blot analysis of M2.Stop(2). 32P-labeled molecular size standards (Lambda DNA-BstEII digest; New England Biolabs, Beverly, MA) were included in each Southern blot analysis. WT, wt MHV68; MR, marker rescue virus.
FIG. 3.
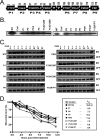
Expression of mutant M2 proteins in mammalian cells. (A) Schematic illustration of the locations of PXXP motifs and tyrosine residues in the M2 protein which have been targeted for mutation in this report. (B) Cos-1 cells were transiently transfected with plasmids expressing the indicated forms of mutant M2 protein. At 48 hours posttransfection, cells were harvested and M2 immunoprecipitations were performed as described in Materials and Methods. (C) Cos-1 cells were transiently transfected with plasmids expressing the indicated forms of mutant M2 protein and GFP. At 24 hours posttransfection, 50 μg/ml CHX (+) or a vehicle control (−) was added to each sample, and cell lysates were harvested at the indicated times, in hours, posttreatment. Protein half-life was calculated by determining the time point at which the band intensity ratio of M2 to GFP was 50% of this ratio at time zero. WT, wt M2; Cntl, vector control; α, anti; IP, immunoprecipitation; _t_1/2, half-life.
FIG. 4.
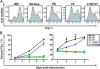
Identification of PXXP motifs and tyrosine residues in the M2 protein that are involved in establishment of and/or reactivation from MHV68 latency in the spleen. C57BL/6J mice were infected intranasally with 100 PFU of wt MHV68, a recombinant M2 mutant virus, or its corresponding marker rescue virus, and spleens were harvested at 16 dpi. Bulk splenocytes were analyzed by limiting-dilution ex vivo reactivation assays and limiting-dilution viral genome PCR assays, as described in Materials and Methods. (A, C, E, G, I, K and M) Frequency of splenocytes harboring viral genome. A limiting-dilution PCR assay was used to determine the frequencies of splenocytes harboring the viral genome. Samples were serially diluted into a background of 104 uninfected cells, lysed, and subjected to nested PCR to detect the viral genome (see Materials and Methods). (B, D, F, H, J, L, and N) Frequency of splenocytes reactivating virus. Results are shown for a limiting-dilution ex vivo reactivation assay in which intact splenocytes were serially diluted on MEF indicator monolayers in parallel with mechanically disrupted samples (to distinguish between virus reactivating from latency and preformed infectious virus). In this report, little or no preformed infectious virus was detected for any of the viruses analyzed (data not shown). For each sample dilution, 24 wells were scored for the presence of CPE (see Materials and Methods). Data are expressed as the mean percentages of wells positive for virus (CPE or viral DNA) ± the standard errors of the means. Curve fit lines for both assays were derived through nonlinear regression analyses. The dashed line indicates 63.2%, from which the frequency of cells reactivating virus or the frequency of cells harboring the viral genome was determined based on Poisson distribution. The data shown represent the means for at least two independent experiments. The recombinant virus nomenclature is described in the legends to Fig. 1 and 3 and Materials and Methods.
FIG. 5.
M2 recombinant viruses display normal acute-phase replication in vivo following intranasal inoculation. C57BL/6J mice were infected intranasally with 100 PFU of a recombinant M2 mutant virus or its corresponding marker rescue virus, and the left lung was extracted at 9 dpi (d9). (A) M2 recombinant viruses with site-directed mutations in PXXP motifs and the corresponding marker rescue viruses. (B) M2 recombinant viruses with site-directed mutations in tyrosine residues and the corresponding marker rescue viruses. The results shown were compiled from two independent experiments with three mice per experiment (each symbol represents data from an individual mouse). Virus titers in lungs were determined by a plaque assay on NIH 3T12 monolayers as described in Materials and Methods. The dashed line indicates the level of detection of the plaque assay (50 PFU), and the solid line indicates the mean virus titer of each group. The recombinant virus nomenclature is described in the legends to Fig. 1 and 3 and Materials and Methods.
FIG. 6.
M2 expression in primary B cells results in expansion of transduced population. (A) Representative flow cytometry data from 3 days posttransduction. (B) M2-expressing B cells expand in culture over time. Triplicate B-cell cultures were transduced with wt M2 or the indicated M2 mutant retrovirus and allowed to rest for 48 h before analysis. Cells were stained with anti-Thy1.1 and analyzed daily for Thy1.1-positive cells. Trypan blue exclusion was used to count absolute numbers of live and dead cells in triplicate wells every day; notably, absolute numbers of cells per well were unchanged over time. Data are representative of two independent experiments.
Similar articles
- Deletion of Murine Gammaherpesvirus Gene M2 in Activation-Induced Cytidine Deaminase-Expressing B Cells Impairs Host Colonization and Viral Reactivation.
Owens SM, Oldenburg DG, White DW, Forrest JC. Owens SM, et al. J Virol. 2020 Dec 9;95(1):e01933-20. doi: 10.1128/JVI.01933-20. Print 2020 Dec 9. J Virol. 2020. PMID: 33028711 Free PMC article. - Disruption of the M2 gene of murine gammaherpesvirus 68 alters splenic latency following intranasal, but not intraperitoneal, inoculation.
Jacoby MA, Virgin HW 4th, Speck SH. Jacoby MA, et al. J Virol. 2002 Feb;76(4):1790-801. doi: 10.1128/jvi.76.4.1790-1801.2002. J Virol. 2002. PMID: 11799175 Free PMC article. - The MHV68 M2 protein drives IL-10 dependent B cell proliferation and differentiation.
Siegel AM, Herskowitz JH, Speck SH. Siegel AM, et al. PLoS Pathog. 2008 Apr 4;4(4):e1000039. doi: 10.1371/journal.ppat.1000039. PLoS Pathog. 2008. PMID: 18389062 Free PMC article. - The murine gammaherpesvirus 68 M2 gene is required for efficient reactivation from latently infected B cells.
Herskowitz JH, Jacoby MA, Speck SH. Herskowitz JH, et al. J Virol. 2005 Feb;79(4):2261-73. doi: 10.1128/JVI.79.4.2261-2273.2005. J Virol. 2005. PMID: 15681428 Free PMC article. - Murine gammaherpesvirus 68 reactivation from B cells requires IRF4 but not XBP-1.
Matar CG, Rangaswamy US, Wakeman BS, Iwakoshi N, Speck SH. Matar CG, et al. J Virol. 2014 Oct;88(19):11600-10. doi: 10.1128/JVI.01876-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078688 Free PMC article.
Cited by
- Themis2/ICB1 is a signaling scaffold that selectively regulates macrophage Toll-like receptor signaling and cytokine production.
Peirce MJ, Brook M, Morrice N, Snelgrove R, Begum S, Lanfrancotti A, Notley C, Hussell T, Cope AP, Wait R. Peirce MJ, et al. PLoS One. 2010 Jul 13;5(7):e11465. doi: 10.1371/journal.pone.0011465. PLoS One. 2010. PMID: 20644716 Free PMC article. - Role of Src homology domain binding in signaling complexes assembled by the murid γ-herpesvirus M2 protein.
Pires de Miranda M, Lopes FB, McVey CE, Bustelo XR, Simas JP. Pires de Miranda M, et al. J Biol Chem. 2013 Feb 8;288(6):3858-70. doi: 10.1074/jbc.M112.439810. Epub 2012 Dec 20. J Biol Chem. 2013. PMID: 23258536 Free PMC article. - Deletion of Murine Gammaherpesvirus Gene M2 in Activation-Induced Cytidine Deaminase-Expressing B Cells Impairs Host Colonization and Viral Reactivation.
Owens SM, Oldenburg DG, White DW, Forrest JC. Owens SM, et al. J Virol. 2020 Dec 9;95(1):e01933-20. doi: 10.1128/JVI.01933-20. Print 2020 Dec 9. J Virol. 2020. PMID: 33028711 Free PMC article. - Tyrosine 129 of the murine gammaherpesvirus M2 protein is critical for M2 function in vivo.
Rangaswamy US, O'Flaherty BM, Speck SH. Rangaswamy US, et al. PLoS One. 2014 Aug 14;9(8):e105197. doi: 10.1371/journal.pone.0105197. eCollection 2014. PLoS One. 2014. PMID: 25122496 Free PMC article. - Characterization of a novel wood mouse virus related to murid herpesvirus 4.
Hughes DJ, Kipar A, Milligan SG, Cunningham C, Sanders M, Quail MA, Rajandream MA, Efstathiou S, Bowden RJ, Chastel C, Bennett M, Sample JT, Barrell B, Davison AJ, Stewart JP. Hughes DJ, et al. J Gen Virol. 2010 Apr;91(Pt 4):867-79. doi: 10.1099/vir.0.017327-0. Epub 2009 Nov 25. J Gen Virol. 2010. PMID: 19940063 Free PMC article.
References
- Babcock, G. J., D. Hochberg, and A. D. Thorley-Lawson. 2000. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity 13497-506. - PubMed
- Beaufils, P., D. Choquet, R. Z. Mamoun, and B. Malissen. 1993. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 125105-5112. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA058524/CA/NCI NIH HHS/United States
- R01 CA087650/CA/NCI NIH HHS/United States
- R01 CA043143/CA/NCI NIH HHS/United States
- R01 AI058057/AI/NIAID NIH HHS/United States
- R01 CA095318/CA/NCI NIH HHS/United States
- R01 CA87650/CA/NCI NIH HHS/United States
- R01 CA58524/CA/NCI NIH HHS/United States
- R01 CA43143/CA/NCI NIH HHS/United States
- R01 AI58057/AI/NIAID NIH HHS/United States
- R01 CA95318/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous