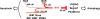Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68 - PubMed (original) (raw)
Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68
Bonsu Ku et al. PLoS Pathog. 2008.
Abstract
All gammaherpesviruses express homologues of antiapoptotic B-cell lymphoma-2 (BCL-2) to counter the clearance of infected cells by host antiviral defense machineries. To gain insights into the action mechanisms of these viral BCL-2 proteins, we carried out structural and biochemical analyses on the interactions of M11, a viral BCL-2 of murine gamma-herpesvirus 68, with a fragment of proautophagic Beclin1 and BCL-2 homology 3 (BH3) domain-containing peptides derived from an array of proapoptotic BCL-2 family proteins. Mainly through hydrophobic interactions, M11 bound the BH3-like domain of Beclin1 with a dissociation constant of 40 nanomole, a markedly tighter affinity compared to the 1.7 micromolar binding affinity between cellular BCL-2 and Beclin1. Consistently, M11 inhibited autophagy more efficiently than BCL-2 in NIH3T3 cells. M11 also interacted tightly with a BH3 domain peptide of BAK and those of the upstream BH3-only proteins BIM, BID, BMF, PUMA, and Noxa, but weakly with that of BAX. These results collectively suggest that M11 potently inhibits Beclin1 in addition to broadly neutralizing the proapoptotic BCL-2 family in a similar but distinctive way from cellular BCL-2, and that the Beclin1-mediated autophagy may be a main target of the virus.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
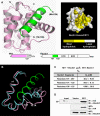
Figure 1. Structural and Binding Analyses of the M11–Beclin1(101–150) Complex
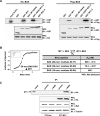
(A) Ribbon drawing (left) and surface presentation (right). M11 is in pink, while the Beclin1 helix is in green. The pink and green regions on the primary sequence diagrams indicate the fragments of the M11 and Beclin1 used for the structure determination. “HT”, “CCD” and “ECD” denote hydrophobic tail, coiled-coil domain and evolutionarily conserved domain, respectively. Only 19 amino acids of Beclin1 exhibited well-defined electron density. A surface presentation of M11 with the omission of the Beclin1 helix shows that the BH3-binding groove is predominantly hydrophobic. The surface coloring scheme is as follows: olive for Val, Leu, Ile, Phe, Trp, Met, and Ala; yellow for Cys, Gly, Tyr, and Pro; gray for other amino acids. (B) Large conformational change of M11 induced by the Beclin1 binding. Only the BH3-binding groove region of M11 is shown for clarity. Beclin1(101–150)-bound M11 (pink) and free M11 (cyan) are superposed. The bound Beclin1 peptide is in green. Helix α3 of M11 undergoes a pronounced conformational change. The arrows indicate the movements of the Cα atoms of Asp59 and Tyr60 in M11. (C) ITC analysis. The measurements were carried out by titrating 0.1 mM of M11 into 5 μM of the indicated Beclin1 fragments. The K D values were deduced from curve fittings of the integrated heat per mol of added ligand and summarized in the table. (D) M11 interacts with endogenous Beclin1. NIH3T3 cells were transfected with HA-tagged M11 and whole cell lysates were used for immunoprecipitation with anti-HA followed by immunoblotting with anti-Beclin1
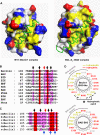
Figure 2. Beclin1 Has a BH3-Like Domain Containing an Atypical Threonine and an Exposed Hydrophobic Patch
(A) A structural comparison of the M11–Beclin1(101–150) (left) and the BCL-XL–BAD complexes (right). M11 and BCL-XL are shown as surface models. The Beclin1 and BAD residues shown in sticks correspond to the five BH3 residues that are critical for the interactions with antiapoptotic BCL-2 family members [21]. They occupy equivalent positions at the BH3-binding groove in the two structures. The surface coloring scheme is as follows: yellow for Val, Leu, Ile, Tyr, Phe, Trp, Met, and Ala; blue for Lys, Arg, and His; red for Glu and Asp; gray for other amino acids. (B) Sequence comparison of the BH3-like domain of mouse Beclin1 with various BH3 domains. Conserved residues are highlighted by red or pink columns. The arrows indicate the five BH3 residues shown in (A). Of these, Thr117 of Beclin1 (red arrow) is not conserved. (C) Sequence alignment. The BH3-like domains of Beclin1 orthologues are aligned (mm, mouse; hs, human; xl, Xenopus laevis; tr, Takifugu rubripes; dm, Drosophila melanogaster; sc, Saccharomyces cerevisiae). The arrows at the top indicate the BH3 residues shown in (A). These residues are highly conserved throughout species, except for Thr117 of mouse Beclin1, which is conserved only in the vertebrates. The conserved hydrophobic residues of Beclin1 exposed in the structure are indicated by the blue arrows at the bottom. (D) α-helical wheel representation. The Beclin1 α-helix bound to M11 is compared with the BAD α-helix bound to BCL-XL. The Beclin1 helix has a hydrophobic patch (indicated by an asterisk) on the opposite side of the BH3-binding groove, unlike the BAD helix.
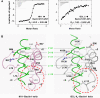
Figure 3. M11 Interacts Much More Tightly with Beclin1 than BCL-2 and BCL-XL
(A) ITC analysis. The measurement was carried out by titrating 0.1 mM of M11 or BCL-2 into 5 μM of the indicated Beclin1 fragment. The K D values were deduced from curve fittings of the integrated heat per mol of added ligand. (B) M11–Beclin1 interface is tighter than that of BCL-XL–Beclin1. The structures of M11–Beclin1 fragment (left) and BCL-XL–Beclin1 peptide (right) are compared side by side. In both the structures, the five consensus BH3 residues of the bound α-helices and the side chains of M11 or BCL-XL interacting with those residues are shown as sticks and labeled. Noted are the tighter interactions of the N-terminal region of Beclin1 with the α3 helix of M11 than the corresponding region in the BCL-XL–Beclin1 helix (indicated by dotted circles).
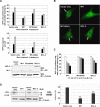
Figure 4. M11 Inhibits Autophagosome Formation in NIH3T3 Cells More Efficiently than BCL-2
(A) Light microscopic quantification of autophagy. After transfection with a GFP–LC3 expression plasmid together with the vector encoding the indicated protein, cells were maintained under normal conditions or treated with 2 μM rapamycin for 4 h. M11(AAA) is an M11 mutant containing three alanine substitutions at the BH3-binding groove. Autophagy was quantified as the percentage of GFP–LC3 positive cells (top) or as the number of autophagosomes (GFP–LC3 positive dots) per cell (bottom). The expression of M11 resulted in fewer GFP–LC3 positive cells or spots than the expression of BCL-2. Data represent mean ± s.d. of three experiments. Expression levels of M11, M11(AAA) and BCL-2 are shown below. (B) Confocal microscopic images of the rafamycin treated cells. GFP–LC3 was detected using an inverted fluorescence microscope. Arrows indicate autophagosomes labeled with GFP–LC3. (C) Dose response. NIH3T3 cells were transfected with GFP–LC3 expression plasmid together with increasing amount of plasmid encoding the indicated proteins. At 16–18 h posttransfection, cells were subjected to 2 μM rapamycin treatment for 4 h and autophagy level was quantified as described at (A). (D) LC3 mobility shift. The whole cells lysates of the rafamycin treated cells were subjected to immunoblotting with anti-LC3 and anti-tubulin antibodies (left). The ratio of quantified band intensities is also shown (right). The cleaved form (LC3-II) of the LC3 precursor (LC3-I) was undetectable and the level of LC3-II/LC3-I was far lower in the cells expressing M11 in contrast with the cells expressing BCL-2. Data represent mean ± s.d. of three experiments. **, P < 0.005 versus vector (Student t test).
Figure 5. Analyses of the Interactions between M11 or BCL-2 Proteins and BAX/BAK
(A) Cell-based binding assay. 293T cells were transfected with HA-tagged BAK or Flag-tagged BAX together with the indicated GST-tagged prosurvival BCL-2 proteins. Whole cell lysates were used for immunoprecipitation with anti-GST followed by immunoblotting with anti-HA or anti-Flag. While no band was detected for the interaction of GST–KSHV BCL-2 with Flag–BAX in this run, a faint band was detected in another run (Figure S5). (B) ITC analyses of the interactions of M11 with the BH3 peptides of BAX or BAK. The ITC analysis was carried out by titrating 0.1 mM of the indicated peptides into 5 μM of M11. The ITC run for the titration of the 26-mer BAK peptide is shown. The deduced K D values are shown in the table. (C) M11 interacts with endogenous BAK. NIH3T3 cells were transfected with HA-tagged M11 and whole cell lysates were used for immunoprecipitation with control rabbit serum or anti-BAK followed by immunoblotting with anti-HA.
Figure 6. ITC Analyses of the Interactions of M11 with the BH3 Peptides of BH3-Only Proteins
Representative ITC runs for the interaction of M11 with the Noxa and BAD peptides are shown, and the K D values determined by this method are summarized in the table.
Figure 7. Model for M11 Action Mechanism
The binding analyses presented in this study suggest that M11 antagonizes cell death by simultaneous inhibition of apoptosis and autophagy. The varied thickness of the arrows denoting the negative regulation by M11 indicates the inhibitory potency according to the K D values determined in this study and shown next to the arrows. Dashed line is used to indicate that BAX may be inhibited by M11 but only weakly. Although not indicated in the figure, cellular BCL-2 proteins, protected by M11, may sequester and inhibit BAX (see text). PI(3)KCIII and UVRAG stand for class III phosphatidylinositol 3-kinase and UV irradiation resistance-associated gene, respectively.
Similar articles
- Targeting γ-herpesvirus 68 Bcl-2-mediated down-regulation of autophagy.
Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S. Su M, et al. J Biol Chem. 2014 Mar 21;289(12):8029-40. doi: 10.1074/jbc.M113.515361. Epub 2014 Jan 17. J Biol Chem. 2014. PMID: 24443581 Free PMC article. - Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11.
Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Sinha S, et al. Autophagy. 2008 Nov;4(8):989-97. doi: 10.4161/auto.6803. Epub 2008 Nov 18. Autophagy. 2008. PMID: 18797192 Free PMC article. - Reconstitution of interactions of Murine gammaherpesvirus 68 M11 with Bcl-2 family proteins in yeast.
Juhásová B, Bhatia-Kiššová I, Polčicová K, Mentel M, Forte M, Polčic P. Juhásová B, et al. Biochem Biophys Res Commun. 2011 Apr 22;407(4):783-7. doi: 10.1016/j.bbrc.2011.03.100. Epub 2011 Mar 31. Biochem Biophys Res Commun. 2011. PMID: 21439939 - Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.
Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Salminen A, et al. Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review. - The autophagy effector Beclin 1: a novel BH3-only protein.
Sinha S, Levine B. Sinha S, et al. Oncogene. 2008 Dec;27 Suppl 1(Suppl 1):S137-48. doi: 10.1038/onc.2009.51. Oncogene. 2008. PMID: 19641499 Free PMC article. Review.
Cited by
- Locating Herpesvirus Bcl-2 Homologs in the Specificity Landscape of Anti-Apoptotic Bcl-2 Proteins.
Foight GW, Keating AE. Foight GW, et al. J Mol Biol. 2015 Jul 31;427(15):2468-2490. doi: 10.1016/j.jmb.2015.05.015. Epub 2015 May 23. J Mol Biol. 2015. PMID: 26009469 Free PMC article. - Viral interactions with macroautophagy: a double-edged sword.
Lin LT, Dawson PW, Richardson CD. Lin LT, et al. Virology. 2010 Jun 20;402(1):1-10. doi: 10.1016/j.virol.2010.03.026. Epub 2010 Apr 21. Virology. 2010. PMID: 20413139 Free PMC article. Review. - An integrated approach to elucidate the intra-viral and viral-cellular protein interaction networks of a gamma-herpesvirus.
Lee S, Salwinski L, Zhang C, Chu D, Sampankanpanich C, Reyes NA, Vangeloff A, Xing F, Li X, Wu TT, Sahasrabudhe S, Deng H, Lacount DJ, Sun R. Lee S, et al. PLoS Pathog. 2011 Oct;7(10):e1002297. doi: 10.1371/journal.ppat.1002297. Epub 2011 Oct 20. PLoS Pathog. 2011. PMID: 22028648 Free PMC article. - Autophagy regulation in cancer development and therapy.
White EJ, Martin V, Liu JL, Klein SR, Piya S, Gomez-Manzano C, Fueyo J, Jiang H. White EJ, et al. Am J Cancer Res. 2011;1(3):362-372. Epub 2010 Jan 25. Am J Cancer Res. 2011. PMID: 21969237 Free PMC article. - The Viral Bcl-2 Homologs of Kaposi's Sarcoma-Associated Herpesvirus and Rhesus Rhadinovirus Share an Essential Role for Viral Replication.
Gallo A, Lampe M, Günther T, Brune W. Gallo A, et al. J Virol. 2017 Feb 28;91(6):e01875-16. doi: 10.1128/JVI.01875-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28053098 Free PMC article.
References
- Cuconati A, White E. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 2002;16:2465–2478. - PubMed
- Hardwick JM, Bellows DS. Viral versus cellular BCL-2 proteins. Cell Death Differ. 2003;10:S68–S76. - PubMed
- Meseda CA, Arrand JR, Mackett M. Herpesvirus papio encodes a functional homologue of the Epstein-Barr virus apoptosis suppressor, BHRF1. J Gen Virol. 2000;81:1801–1805. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials