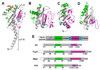Type IV pili: paradoxes in form and function - PubMed (original) (raw)
Review
Type IV pili: paradoxes in form and function
Lisa Craig et al. Curr Opin Struct Biol. 2008 Apr.
Abstract
Type IV pili are filaments on the surfaces of many Gram-negative bacteria that mediate an extraordinary array of functions, including adhesion, motility, microcolony formation and secretion of proteases and colonization factors. Their prominent display on the surfaces of many bacterial pathogens, their vital role in virulence, and their ability to elicit an immune response make Type IV pilus structures particularly relevant for study as targets for component vaccines and therapies. Structural studies of the pili and components of the pilus assembly apparatus have proven extremely challenging, but new approaches and methods have produced important breakthroughs that are advancing our understanding of pilus functions and their complex assembly mechanism. These structures provide insights into the biology of Type IV pili as well as that of the related bacterial secretion and archaeal flagellar systems. This review will summarize the most recent structural advances on Type IV pili and their assembly components and highlight their significance.
Figures
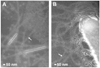
Fig. 1. Neisseria gonorrhoeae GC pili and Vibrio cholerae toxin-coregulated pili imaged by negative-stain electron microscopy
(A) GC pili are indicated by arrows. Tobacco mosaic virus particles (18 Å diameter) are also present. (B) TCP (arrows) emanate from the V. cholerae surface.
Fig. 2. Structure and schematic representations of Type IV pilin subunits and the pilin-like protein, PilX
X-ray crystal structures of (A) full length N. gonorrhoeae GC pilin at 2.3 Å resolution [8] and (B) N-terminally truncated V. cholerae TcpA at 1.3 Å resolution [10]. GC pilin has two post-translational modifications: a disaccharide α-D-galactopyranosyl-(1→3)-2,4-diacetamido-2,4-dideoxy-β-D-glucopyranoside covalently attached to Ser63 and a phosphoethanolamine at Ser68. (C) NMR structure of N-terminally truncated EPEC BfpA [11]. (D) X-ray crystal structure of N-terminally truncated N. meningitidis PilX at 2.4 Å resolution [15]. The αβ-loops are colored green and the D-regions are colored magenta. The disulfide-bonded cysteines are shown in yellow and cyan. (E) Schematic representation of the pilins and PilX indicating the relevant regions and residues. The jagged line in TcpA, BfpA and PilX represents the site of truncation for structure determination.
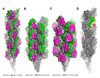
Fig. 3. Type IV pilus models
(A) CryoEM reconstruction of the N. gonorrhoeae GC pilus at 12.5 Å resolution colored as in Fig. 2 [8]. (B) EM-based model of EPEC BFP [11]. (C) V. cholerae TCP model based on DXMS analysis and EM-derived structural parameters [12]. Arrows indicate the exposed segment of α1-N. (D) GC pilus model with two subunits replaced by N. meningitidis PilX, colored yellow with the αβ-loop and D-region colored green and magenta, respectively.
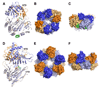
Fig. 4. Crystal structures of A. aeolicus PilT and A. fulgidus GspE2 ATPases
(A) Subunits E (orange) and F (blue) of the 4.2 Å PilT structure [28], representing the open and closed states, respectively, and superimposed via the C-terminal domains (CTD). Dark colors are used for the N-terminal domains (NTD) and light colors are used for the CTD. The NTD arginine fingers are shown as sticks and bound ADP is shown in green and orange ball-and-sticks. The AIRNLIRE motif α-helix (see text) is colored green at the bottom of the CTD. (B) End view of the asymmetric PilT hexamer viewed from the NTD side. (C) Side view of the PilT hexamer. (D) Superposition of the CTDs of the open (orange) and closed (blue) forms of GspE2 bound to AMP-PNP at 2.95 Å resolution [29]. (E) End view and (F) side view of the GspE2 hexamer with alternating open and closed conformations.
Fig. 5. EM reconstruction of the N. meningitidis inner membrane protein PilG
Side and top views of a four-fold symmetrized PilG reconstruction from negatively-stained EM images [33]. The shaded region represents the putative transmembrane “waist”. Nanogold particles bound to the cone-shaped lower domain, indicating the N-terminal region, which is predicted to be located on the cytoplasmic side of the inner membrane.
Fig. 6. Model for Type IV pilus assembly
In Step 1, pilin subunits diffuse throughout the inner membrane (IM) and encounter (or are recruited to) the pilus assembly apparatus. The negative charge on Glu5, which makes the subunits somewhat unstable in the lipid bilayer, is attracted to the positively-charged N-terminus of the most terminal pilin subunit in the growing filament. Additional attractive forces between the globular domains allow the subunit to dock into an existing gap at the filament base, thus adding one subunit to the 3-start helical strand colored in red (Step 2). The assembly ATPase is associated with the cytoplasmic side of the inner membrane, possibly via an integral membrane protein (IMP). ATP is bound in the active site cleft of one of the assembly ATPase subunits, causing the N-terminal domain to clamp down on the C-terminal domain. In Step 3, the ATP is hydrolyzed, releasing the N-terminal domain, which twists away from the C-terminal domain. This domain movement induces a conformational change or shift in the associated IMP, which forces the pilus filament out of the membrane by a short distance (~10 Å). This movement results in a new gap at the next strand of the 3-start helix, ready for the addition of a new pilin subunit.
Fig. 7. Structures of the N. meningitidis PilQ secretin complex and PilP lipoprotein
(A) CryoEM reconstruction of the PilQ complex at 12 Å resolution [39]. (B) NMR structure of the PilP lipoprotein fragment [43].
Similar articles
- The Biosynthesis and Structures of Bacterial Pili.
Lukaszczyk M, Pradhan B, Remaut H. Lukaszczyk M, et al. Subcell Biochem. 2019;92:369-413. doi: 10.1007/978-3-030-18768-2_12. Subcell Biochem. 2019. PMID: 31214993 Review. - Pili in Gram-negative and Gram-positive bacteria - structure, assembly and their role in disease.
Proft T, Baker EN. Proft T, et al. Cell Mol Life Sci. 2009 Feb;66(4):613-35. doi: 10.1007/s00018-008-8477-4. Cell Mol Life Sci. 2009. PMID: 18953686 Free PMC article. Review. - Towards a systems biology approach to study type II/IV secretion systems.
Hazes B, Frost L. Hazes B, et al. Biochim Biophys Acta. 2008 Sep;1778(9):1839-50. doi: 10.1016/j.bbamem.2008.03.011. Epub 2008 Mar 21. Biochim Biophys Acta. 2008. PMID: 18406342 Review. - A comprehensive guide to pilus biogenesis in Gram-negative bacteria.
Hospenthal MK, Costa TRD, Waksman G. Hospenthal MK, et al. Nat Rev Microbiol. 2017 May 12;15(6):365-379. doi: 10.1038/nrmicro.2017.40. Nat Rev Microbiol. 2017. PMID: 28496159 Review. - Chemical attenuation of pilus function and assembly in Gram-negative bacteria.
Lo AW, Moonens K, Remaut H. Lo AW, et al. Curr Opin Microbiol. 2013 Feb;16(1):85-92. doi: 10.1016/j.mib.2013.02.003. Epub 2013 Feb 19. Curr Opin Microbiol. 2013. PMID: 23434114 Review.
Cited by
- Genetic and mass spectrometry analyses of the unusual type IV-like pili of the archaeon Methanococcus maripaludis.
Ng SY, Wu J, Nair DB, Logan SM, Robotham A, Tessier L, Kelly JF, Uchida K, Aizawa S, Jarrell KF. Ng SY, et al. J Bacteriol. 2011 Feb;193(4):804-14. doi: 10.1128/JB.00822-10. Epub 2010 Nov 12. J Bacteriol. 2011. PMID: 21075925 Free PMC article. - Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential.
Hartung S, Arvai AS, Wood T, Kolappan S, Shin DS, Craig L, Tainer JA. Hartung S, et al. J Biol Chem. 2011 Dec 23;286(51):44254-44265. doi: 10.1074/jbc.M111.297242. Epub 2011 Oct 24. J Biol Chem. 2011. PMID: 22027840 Free PMC article. - More than one way to control hair growth: regulatory mechanisms in enterobacteria that affect fimbriae assembled by the chaperone/usher pathway.
Clegg S, Wilson J, Johnson J. Clegg S, et al. J Bacteriol. 2011 May;193(9):2081-8. doi: 10.1128/JB.00071-11. Epub 2011 Mar 11. J Bacteriol. 2011. PMID: 21398554 Free PMC article. Review. - Type IV pili in Gram-positive bacteria.
Melville S, Craig L. Melville S, et al. Microbiol Mol Biol Rev. 2013 Sep;77(3):323-41. doi: 10.1128/MMBR.00063-12. Microbiol Mol Biol Rev. 2013. PMID: 24006467 Free PMC article. Review. - The Flp type IV pilus operon of Mycobacterium tuberculosis is expressed upon interaction with macrophages and alveolar epithelial cells.
Alteri CJ, Rios-Sarabia N, De la Cruz MA, González-Y-Merchand JA, Soria-Bustos J, Maldonado-Bernal C, Cedillo ML, Yáñez-Santos JA, Martínez-Laguna Y, Torres J, Friedman RL, Girón JA, Ares MA. Alteri CJ, et al. Front Cell Infect Microbiol. 2022 Sep 20;12:916247. doi: 10.3389/fcimb.2022.916247. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36204636 Free PMC article.
References
- Remaut H, Waksman G. Structural biology of bacterial pathogenesis. Curr Opin Struct Biol. 2004;14:161–170. - PubMed
- Scott JR, Zahner D: Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. - PubMed
- Bardy SL, Ng SY, Jarrell KF. Prokaryotic motility structures. Microbiology. 2003;149:295–304. - PubMed
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. - PubMed
- Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS Microbiol Lett. 2006;255:175–186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR001209/RR/NCRR NIH HHS/United States
- P41 RR001209-277563/RR/NCRR NIH HHS/United States
- R21 AI061051/AI/NIAID NIH HHS/United States
- R21 AI061051-03/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources