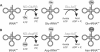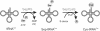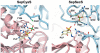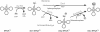From one amino acid to another: tRNA-dependent amino acid biosynthesis - PubMed (original) (raw)
Review
. 2008 Apr;36(6):1813-25.
doi: 10.1093/nar/gkn015. Epub 2008 Feb 5.
Affiliations
- PMID: 18252769
- PMCID: PMC2330236
- DOI: 10.1093/nar/gkn015
Review
From one amino acid to another: tRNA-dependent amino acid biosynthesis
Kelly Sheppard et al. Nucleic Acids Res. 2008 Apr.
Abstract
Aminoacyl-tRNAs (aa-tRNAs) are the essential substrates for translation. Most aa-tRNAs are formed by direct aminoacylation of tRNA catalyzed by aminoacyl-tRNA synthetases. However, a smaller number of aa-tRNAs (Asn-tRNA, Gln-tRNA, Cys-tRNA and Sec-tRNA) are made by synthesizing the amino acid on the tRNA by first attaching a non-cognate amino acid to the tRNA, which is then converted to the cognate one catalyzed by tRNA-dependent modifying enzymes. Asn-tRNA or Gln-tRNA formation in most prokaryotes requires amidation of Asp-tRNA or Glu-tRNA by amidotransferases that couple an amidase or an asparaginase to liberate ammonia with a tRNA-dependent kinase. Both archaeal and eukaryotic Sec-tRNA biosynthesis and Cys-tRNA synthesis in methanogens require O-phosophoseryl-tRNA formation. For tRNA-dependent Cys biosynthesis, O-phosphoseryl-tRNA synthetase directly attaches the amino acid to the tRNA which is then converted to Cys by Sep-tRNA: Cys-tRNA synthase. In Sec-tRNA synthesis, O-phosphoseryl-tRNA kinase phosphorylates Ser-tRNA to form the intermediate which is then modified to Sec-tRNA by Sep-tRNA:Sec-tRNA synthase. Complex formation between enzymes in the same pathway may protect the fidelity of protein synthesis. How these tRNA-dependent amino acid biosynthetic routes are integrated into overall metabolism may explain why they are still retained in so many organisms.
Figures
Figure 1.
Indirect pathways for (A) Gln-tRNAGln and (B) Asn-tRNAAsn formation. (A) First a ND-GluRS glutamylates tRNAGln to form Glu-tRNAGln. The mischarged species is then amidated by a Glu-AdT to form Gln-tRNAGln. (B) First a ND-AspRS aspartylates tRNAAsn to form Asp-tRNAAsn. The mischarged species is then amidated by an Asp-AdT for form Asn-tRNAAsn.
Figure 2.
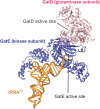
Crystal structure of the M. thermautotrophicus GatDE complexed with tRNAGln. The AdT forms an α2β2 tetramer, with two GatE subunits binding a GatD homodimer. Each GatE subunit binds one tRNAGln molecule. For clarity only one monomer of GatD and GatE are shown. The glutaminase active site of the D-subunit and the kinase active site of the E-subunit are connected by a 40 Å long molecular tunnel (44). Adapted from Polycarpo,C. et al. (2007). In Cavicchioli,R. (ed.) Archaea: Molecular and Cellular Biology. ASM Press, Washington, DC USA with permission from ASM Press.
Figure 3.
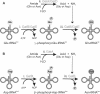
Both GatCAB (A and B) and GatDE (A) catalyze three distinct reactions in order to transamidate their mischarged tRNA species, (A) Glu-tRNAGln and/or (B) Asp-tRNAAsn: (i) the kinase subunit of the respective AdT (GatB or GatE) phosphorylates the mischarged tRNA species to form an activated intermediate, (A) γ-phosphoryl-Glu-tRNAGln or (B) β-phosphoryl-Asp-tRNAAsn; (ii) the glutaminase subunit (GatA or GatD) hydrolyzes an amide donor such as Gln or Asn to release ammonia. A molecular tunnel connects the glutaminase and kinase active sites of the respective AdTs, allowing ammonia liberated from the glutamianse subunit (GatA or GatD) to flow to the kinase subunit (GatB or GatE) (denoted by the dashed arrow); (iii) the liberated ammonia is then used by the kinase subunit (GatB or GatE) to amidate the activated intermediate to form the product aa-tRNA, (A) Gln-tRNAGln or (B) Asn-tRNAAsn.
Figure 4.
Indirect pathway for Cys-tRNACys formation. First, SepRS aminoacylates tRNACys with Sep to form Sep-tRNACys. The Sep bound to the tRNA is then converted to Cys by SepCysS in the presence of a sulfur donor to form Cys-tRNACys.
Figure 5.
The crystal structures of the active sites of A. fulgidus SepCysS and M. maripaludis SepSecS. In both, the different monomers of the respective enzyme are colored pink and blue. PLP and residues in the catalytic centers are shown as ball-and-stick models (adapted from 136).
Figure 6.
Indirect pathways for Sec-tRNASec formation. In all known Sec-decoding organisms, first SerRS aminoacylates tRNASec with Ser to form Ser-tRNASec. In Sec-decoding bacteria, the Ser bound to the tRNA is directly converted to Sec in the presence of selenophosphate by SelA to form Sec-tRNASec. In Sec-decoding eukaryotes and Archaea, the Ser-moiety on tRNASec is first phosphorylated by PSTK to form Sep-tRNASec. The Sep bound to the tRNA is then converted to Sec in the presence of selenophosphate by SepSecS to form Sec-tRNASec.
Similar articles
- Amino acid modifications on tRNA.
Yuan J, Sheppard K, Söll D. Yuan J, et al. Acta Biochim Biophys Sin (Shanghai). 2008 Jul;40(7):539-53. doi: 10.1111/j.1745-7270.2008.00435.x. Acta Biochim Biophys Sin (Shanghai). 2008. PMID: 18604446 Free PMC article. Review. - Aminoacyl-tRNA synthesis by pre-translational amino acid modification.
Feng L, Sheppard K, Namgoong S, Ambrogelly A, Polycarpo C, Randau L, Tumbula-Hansen D, Söll D. Feng L, et al. RNA Biol. 2004 May;1(1):16-20. Epub 2004 May 28. RNA Biol. 2004. PMID: 17194933 Review. - RNA-Dependent Cysteine Biosynthesis in Bacteria and Archaea.
Mukai T, Crnković A, Umehara T, Ivanova NN, Kyrpides NC, Söll D. Mukai T, et al. mBio. 2017 May 9;8(3):e00561-17. doi: 10.1128/mBio.00561-17. mBio. 2017. PMID: 28487430 Free PMC article. - A tRNA-dependent cysteine biosynthesis enzyme recognizes the selenocysteine-specific tRNA in Escherichia coli.
Yuan J, Hohn MJ, Sherrer RL, Palioura S, Su D, Söll D. Yuan J, et al. FEBS Lett. 2010 Jul 2;584(13):2857-61. doi: 10.1016/j.febslet.2010.05.028. Epub 2010 May 21. FEBS Lett. 2010. PMID: 20493852 Free PMC article. - Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis.
Turanov AA, Xu XM, Carlson BA, Yoo MH, Gladyshev VN, Hatfield DL. Turanov AA, et al. Adv Nutr. 2011 Mar;2(2):122-8. doi: 10.3945/an.110.000265. Epub 2011 Mar 10. Adv Nutr. 2011. PMID: 22332041 Free PMC article. Review.
Cited by
- On the Track of the Missing tRNA Genes: A Source of Non-Canonical Functions?
Ehrlich R, Davyt M, López I, Chalar C, Marín M. Ehrlich R, et al. Front Mol Biosci. 2021 Mar 16;8:643701. doi: 10.3389/fmolb.2021.643701. eCollection 2021. Front Mol Biosci. 2021. PMID: 33796548 Free PMC article. Review. - The structural basis of the genetic code: amino acid recognition by aminoacyl-tRNA synthetases.
Kaiser F, Krautwurst S, Salentin S, Haupt VJ, Leberecht C, Bittrich S, Labudde D, Schroeder M. Kaiser F, et al. Sci Rep. 2020 Jul 28;10(1):12647. doi: 10.1038/s41598-020-69100-0. Sci Rep. 2020. PMID: 32724042 Free PMC article. - Non-canonical roles of tRNAs and tRNA mimics in bacterial cell biology.
Katz A, Elgamal S, Rajkovic A, Ibba M. Katz A, et al. Mol Microbiol. 2016 Aug;101(4):545-58. doi: 10.1111/mmi.13419. Epub 2016 Jun 28. Mol Microbiol. 2016. PMID: 27169680 Free PMC article. Review. - Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes.
Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DP, Placha G, Canani LH, Bochenski J, Waggott D, Merchant ML, Krolewski B, Mirea L, Wanic K, Katavetin P, Kure M, Wolkow P, Dunn JS, Smiles A, Walker WH, Boright AP, Bull SB; DCCT/EDIC Research Group; Doria A, Rogus JJ, Rich SS, Warram JH, Krolewski AS. Pezzolesi MG, et al. Diabetes. 2009 Jun;58(6):1403-10. doi: 10.2337/db08-1514. Epub 2009 Feb 27. Diabetes. 2009. PMID: 19252134 Free PMC article. - Yeast mitochondrial Gln-tRNA(Gln) is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS.
Frechin M, Senger B, Brayé M, Kern D, Martin RP, Becker HD. Frechin M, et al. Genes Dev. 2009 May 1;23(9):1119-30. doi: 10.1101/gad.518109. Genes Dev. 2009. PMID: 19417106 Free PMC article.
References
- Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. - PubMed
- Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. - PubMed
- Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J. Biol. Chem. 2007;282:11866–11873. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources