Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure - PubMed (original) (raw)
. 2008 Feb 12;105(6):2111-6.
doi: 10.1073/pnas.0710228105. Epub 2008 Feb 6.
Elizabeth P Murchison, Ruhang Tang, Thomas E Callis, Mariko Tatsuguchi, Zhongliang Deng, Mauricio Rojas, Scott M Hammond, Michael D Schneider, Craig H Selzman, Gerhard Meissner, Cam Patterson, Gregory J Hannon, Da-Zhi Wang
Affiliations
- PMID: 18256189
- PMCID: PMC2542870
- DOI: 10.1073/pnas.0710228105
Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure
Jian-Fu Chen et al. Proc Natl Acad Sci U S A. 2008.
Abstract
Cardiovascular disease is the leading cause of human morbidity and mortality. Dilated cardiomyopathy (DCM) is the most common form of cardiomyopathy associated with heart failure. Here, we report that cardiac-specific knockout of Dicer, a gene encoding a RNase III endonuclease essential for microRNA (miRNA) processing, leads to rapidly progressive DCM, heart failure, and postnatal lethality. Dicer mutant mice show misexpression of cardiac contractile proteins and profound sarcomere disarray. Functional analyses indicate significantly reduced heart rates and decreased fractional shortening of Dicer mutant hearts. Consistent with the role of Dicer in animal hearts, Dicer expression was decreased in end-stage human DCM and failing hearts and, most importantly, a significant increase of Dicer expression was observed in those hearts after left ventricle assist devices were inserted to improve cardiac function. Together, our studies demonstrate essential roles for Dicer in cardiac contraction and indicate that miRNAs play critical roles in normal cardiac function and under pathological conditions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
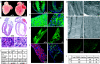
Generation of cardiac-specific Dicer knockout mice. (a) The gene targeting strategy for cardiac-specific deletion of Dicer. (b) PCR genotyping analysis of P0 mice. Wild-type loxP alleles and Cre products are indicated (Upper). Western blot analysis of Dicer protein from wild-type (+/+) and Dicer mutant (−/−) hearts. GAPDH serves as a loading control (Lower). (c) Genotyping results of P0 offspring from MHCCre/+; Dicerflox/+, mice and Dicerflox/flox mice intercrossing. (d) miRNA microarray analysis of miRNA expression in P0 wild-type (+/+), Dicer heterozygous (+/−), and homozygous (−/−) mutant hearts. Red denotes high expression and green denotes low expression. (e and f) miRNA Northern blot analysis to detect the expression of precursor (Pre-) and mature miR-1 and -133 in P0 or embryonic wild-type (+/+) and Dicer mutant (−/−) hearts. tRNAs were used as a loading control. (g) Kaplan–Meier survival curves for wild-type (Wt) and Dicer knockout (Ko) mice. All Dicer mutant mice die before P4.
Fig. 2.
Cardiac-specific Dicer deletion leads to DCM. (a) Gross morphology of P0 wild-type (+/+) and Dicer mutant (−/−) hearts. (b) H&E staining of sagittal sections of P0 wild-type (+/+) and Dicer mutant (−/−) hearts. (c) Higher-magnification images from H&E stained left ventricular myocardium of P0 wild-type (+/+) and Dicer mutant (−/−) hearts. (d) Immunohistology of P0 wild-type (+/+) and Dicer mutant (−/−) hearts for cTnT (green). DAPI stains nuclei. (e) Higher-magnification images from cTnT immunohistology-stained left ventricular myocardium of P0 wild-type (+/+) and Dicer mutant (−/−) hearts. (f) Confocal microscopic images of cultured neonatal cardiomyocytes from wild-type (+/+) and Dicer mutant (−/−) hearts stained for cardiac α-TM. (g) Electron microscopic analysis of ventricular myocardium of wild-type (+/+) and Dicer mutant (−/−) hearts. There is substantially less organized myofibril in Dicer mutant hearts. (h) Higher magnification of electron micrographies of wild-type (+/+) and Dicer mutant (−/−) myocardium to show disarrayed sarcomere in mutant hearts. (i) Quantitative measurement of sarcomere length in wild-type (+/+) and Dicer mutant (−/−) hearts. (j) TUNEL staining (green) detected increased apoptosis in the left atrium and ventricle of P2 Dicer mutant (−/−) heart. Antibody against cTnT was used to label cardiomyocytes (red). DAPI stains nuclei (blue). (k) Increased apoptosis in the left ventricular wall of P2 Dicer mutant (−/−) hearts as revealed by TUNEL staining. (l) Quantitative measurement of TUNEL positive cells in P2 wild-type (+/+) and Dicer mutant (−/−) hearts.
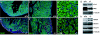
Fig. 3.
Impaired cardiac function in Dicer mutant hearts. (a) M-mode echocardiograms of P0 wild-type (+/+) and Dicer mutant (−/−) mice. (b) Diagram to show the areas of ventricles examined in confocal immunohistochemistry images: area 1 for c and f; area 2 for d and g; area 3 for e and h. (c–h) Confocal microscopic images for Cx40 protein expression (red) in the ventricles of P0 wild-type (+/+) and Dicer mutant (−/−) mice. DAPI stains nuclei. (i) Western blot analyses of connexin protein expression in P0 wild-type (+/+) and Dicer mutant (−/−) mice. β-Tubulin serves as a loading control. (j and k) Confocal microscopic images of Cx45 in P0 wild-type (+/+) and Dicer mutant (−/−) hearts. Laminin marks the cell surface and DAPI stains nuclei.
Fig. 4.
Expression of cardiac contractile proteins in Dicer mutant hearts. (a–d) Confocal microscopic images of P0 wild-type (+/+) and Dicer mutant (−/−) hearts stained with MHC in apexes (a and b) or left ventricle (c and d). Note patches of myocardium that lack expression of MHC in Dicer mutant (−/−) hearts. DAPI stains nuclei. (e and f) Confocal microscopic images of P0 wild-type (+/+) and Dicer mutant (−/−) hearts stained with antibody against α-TM. Note dramatic decrease in the expression of α-TM in Dicer mutant (−/−) hearts. DAPI stains nuclei. (g and h) Western blot analyses of indicated proteins using protein extracts from P0 wild-type (+/+) and Dicer mutant (−/−) hearts. β-tubulin serves as a loading control.
Fig. 5.
Dicer protein expression in patients with failure or nonfailure hearts. Western blot analyses for Dicer from total protein extracts of ventricles of human subjects with end-stage heart failure before (lanes 1–4) or after (lanes 5–8) the application of LVAD, or nonfailure heart samples (lanes 9–11). GAPDH serves as a loading control.
Similar articles
- 4-Hydroxynonenal impairs miRNA maturation in heart failure via Dicer post-translational modification.
Kiyuna LA, Candido DS, Bechara LRG, Jesus ICG, Ramalho LS, Krum B, Albuquerque RP, Campos JC, Bozi LHM, Zambelli VO, Alves AN, Campolo N, Mastrogiovanni M, Bartesaghi S, Leyva A, Durán R, Radi R, Arantes GM, Cunha-Neto E, Mori MA, Chen CH, Yang W, Mochly-Rosen D, MacRae IJ, Ferreira LRP, Ferreira JCB. Kiyuna LA, et al. Eur Heart J. 2023 Nov 21;44(44):4696-4712. doi: 10.1093/eurheartj/ehad662. Eur Heart J. 2023. PMID: 37944136 - Dilated cardiomyopathy mutant tropomyosin mice develop cardiac dysfunction with significantly decreased fractional shortening and myofilament calcium sensitivity.
Rajan S, Ahmed RP, Jagatheesan G, Petrashevskaya N, Boivin GP, Urboniene D, Arteaga GM, Wolska BM, Solaro RJ, Liggett SB, Wieczorek DF. Rajan S, et al. Circ Res. 2007 Jul 20;101(2):205-14. doi: 10.1161/CIRCRESAHA.107.148379. Epub 2007 Jun 7. Circ Res. 2007. PMID: 17556658 - Increased expression of constitutive nitric oxide synthase III, but not inducible nitric oxide synthase II, in human heart failure.
Stein B, Eschenhagen T, Rüdiger J, Scholz H, Förstermann U, Gath I. Stein B, et al. J Am Coll Cardiol. 1998 Nov;32(5):1179-86. doi: 10.1016/s0735-1097(98)00399-4. J Am Coll Cardiol. 1998. PMID: 9809923 - Inhibition of miR-208b improves cardiac function in titin-based dilated cardiomyopathy.
Zhou Q, Schötterl S, Backes D, Brunner E, Hahn JK, Ionesi E, Aidery P, Sticht C, Labeit S, Kandolf R, Gawaz M, Gramlich M. Zhou Q, et al. Int J Cardiol. 2017 Mar 1;230:634-641. doi: 10.1016/j.ijcard.2016.12.171. Epub 2016 Dec 28. Int J Cardiol. 2017. PMID: 28065693 - Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts.
Makarenko I, Opitz CA, Leake MC, Neagoe C, Kulke M, Gwathmey JK, del Monte F, Hajjar RJ, Linke WA. Makarenko I, et al. Circ Res. 2004 Oct 1;95(7):708-16. doi: 10.1161/01.RES.0000143901.37063.2f. Epub 2004 Sep 2. Circ Res. 2004. PMID: 15345656
Cited by
- In vitro quantification of specific microRNA using molecular beacons.
Baker MB, Bao G, Searles CD. Baker MB, et al. Nucleic Acids Res. 2012 Jan;40(2):e13. doi: 10.1093/nar/gkr1016. Epub 2011 Nov 21. Nucleic Acids Res. 2012. PMID: 22110035 Free PMC article. - Differential expression of dicer, miRNAs, and inflammatory markers in diabetic Ins2+/- Akita hearts.
Chavali V, Tyagi SC, Mishra PK. Chavali V, et al. Cell Biochem Biophys. 2014 Jan;68(1):25-35. doi: 10.1007/s12013-013-9679-4. Cell Biochem Biophys. 2014. PMID: 23797610 Free PMC article. - The Abl1 tyrosine kinase is a key player in doxorubicin-induced cardiomyopathy and its p53/p73 cell death mediated signaling differs in atrial and ventricular cardiomyocytes.
Borlak J, Ciribilli Y, Bisio A, Selvaraj S, Inga A, Oh JH, Spanel R. Borlak J, et al. J Transl Med. 2024 Sep 16;22(1):845. doi: 10.1186/s12967-024-05623-8. J Transl Med. 2024. PMID: 39285385 Free PMC article. - FMSM: a novel computational model for predicting potential miRNA biomarkers for various human diseases.
Sun Y, Zhu Z, You ZH, Zeng Z, Huang ZA, Huang YA. Sun Y, et al. BMC Syst Biol. 2018 Dec 31;12(Suppl 9):121. doi: 10.1186/s12918-018-0664-9. BMC Syst Biol. 2018. PMID: 30598090 Free PMC article. - Restoring Ravaged Heart: Molecular Mechanisms and Clinical Application of miRNA in Heart Regeneration.
Shah V, Shah J. Shah V, et al. Front Cardiovasc Med. 2022 Feb 10;9:835138. doi: 10.3389/fcvm.2022.835138. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35224063 Free PMC article. Review.
References
- Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. - PubMed
- Olson EN. A decade of discoveries in cardiac biology. Nat Med. 2004;10:467–474. - PubMed
- Kamisago M, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–1696. - PubMed
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases