The dynamic nature of eukaryotic genomes - PubMed (original) (raw)
The dynamic nature of eukaryotic genomes
Laura Wegener Parfrey et al. Mol Biol Evol. 2008 Apr.
Abstract
Analyses of diverse eukaryotes reveal that genomes are dynamic, sometimes dramatically so. In numerous lineages across the eukaryotic tree of life, DNA content varies within individuals throughout life cycles and among individuals within species. Discovery of examples of genome dynamism is accelerating as genome sequences are completed from diverse eukaryotes. Though much is known about genomes in animals, fungi, and plants, these lineages represent only 3 of the 60-200 lineages of eukaryotes. Here, we discuss diverse genomic strategies in exemplar eukaryotic lineages, including numerous microbial eukaryotes, to reveal dramatic variation that challenges established views of genome evolution. For example, in the life cycle of some members of the "radiolaria," ploidy increases from haploid (N) to approximately 1,000N, whereas intrapopulation variability of the enteric parasite Entamoeba ranges from 4N to 40N. Variation has also been found within our own species, with substantial differences in both gene content and chromosome lengths between individuals. Data on the dynamic nature of genomes shift the perception of the genome from being fixed and characteristic of a species (typological) to plastic due to variation within and between species.
Figures
Figure 1. Distribution of genomic features
Dynamic genomes are widespread across the eukaryotic tree of life. Occurrence of three metrics of genome dynamism are plotted onto our cartoon of the eukaryotic tree of life, the topology of which is derived from our interpretation of multigene genealogies (Parfrey et al. 2006; Rodriguez-Ezpeleta et al. 2007; Yoon et al. Submitted). (p) indicates paraphyly in radiolaria and green algae. Symbols indicate that the feature is reported in at least one taxon within the lineage. ⇑: Somatic polyploidy, ↑↓: Cyclic polyploidy, ◆: Intraspecific genome variation.
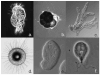
Figure 2. Exemplar microbial eukaryotes
Images of microbial organisms discussed reveal morphological diversity in addition to genomic diversity described in the text. Approximate size is give for reference. (a) Uronychia (ciliate) – 150μm, (b) Ammonia (Foraminifera) – 300 μm, (c) Amoeba proteus – 300 μm, (d) Aulacantha (Phaeodarea) – 200 μm, (e) Entamoeba – 25 μm, (f) Giardia – 12 μm. All except d are images of live organisms, d is a drawing from Haeckel (1862) from the library of Kurt Stueber (
http://caliban.mpiz-koeln.mpg.de/\~stueber/haeckel/radiolarien
). All images used with permission from micro*scope (
http://starcentral.mbl.edu/microscope/portal.php
).
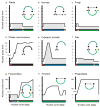
Figure 3. Diversity of nuclear cycles
Depiction of changes in DNA content through the nuclear cycles of nine lineages of eukaryotes. Horizontal axis and colors corresponds to the nuclear cycle stage as shown in the inset diagram, while the vertical axis measure approximate DNA content within the nucleus. Gray shading represents periods of multicellularity or multinuclearity. Inset diagram in panel a is the generalized nuclear cycle as exemplified by plants. Arrows represent progression of genome through karyogamy (red), mitosis as a diploid (blue), meiosis (yellow), and mitosis as a haploid (green). Amitosis in ciliates is black. The nuclear cycle of organisms may include some or all components of the generalized nuclear cycle. A dashed arrow indicates the absence of intervening steps. Panels d and e depict the fate of the somatic genomes, therefore they are dead ends.
Figure 4. Range of intraspecific variation
Individuals within a species (and population) do not have identical genomes. Intraspecific genomes can be different from the level of a few nucleotides, to chromosomes, to ploidy of the whole genome. We plot examples of this variation discussed in the text along a gradient of variation. The number of nucleotides involved increases from left to right.
Similar articles
- Foraminifera as a model of eukaryotic genome dynamism.
Timmons C, Le K, Rappaport HB, Sterner EG, Maurer-Alcalá XX, Goldstein ST, Katz LA. Timmons C, et al. mBio. 2024 Mar 13;15(3):e0337923. doi: 10.1128/mbio.03379-23. Epub 2024 Feb 8. mBio. 2024. PMID: 38329358 Free PMC article. - The dynamic nature of genomes across the tree of life.
Oliverio AM, Katz LA. Oliverio AM, et al. Genome Biol Evol. 2014 Mar;6(3):482-8. doi: 10.1093/gbe/evu024. Genome Biol Evol. 2014. PMID: 24500971 Free PMC article. - Evolutionary convergence on highly-conserved 3' intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome.
Irimia M, Roy SW. Irimia M, et al. PLoS Genet. 2008 Aug 8;4(8):e1000148. doi: 10.1371/journal.pgen.1000148. PLoS Genet. 2008. PMID: 18688272 Free PMC article. - An epigenetic toolkit allows for diverse genome architectures in eukaryotes.
Maurer-Alcalá XX, Katz LA. Maurer-Alcalá XX, et al. Curr Opin Genet Dev. 2015 Dec;35:93-9. doi: 10.1016/j.gde.2015.10.005. Epub 2015 Nov 30. Curr Opin Genet Dev. 2015. PMID: 26649755 Free PMC article. Review. - Transposable elements and factors influencing their success in eukaryotes.
Pritham EJ. Pritham EJ. J Hered. 2009 Sep-Oct;100(5):648-55. doi: 10.1093/jhered/esp065. Epub 2009 Aug 7. J Hered. 2009. PMID: 19666747 Free PMC article. Review.
Cited by
- Protein Bioinformatics Infrastructure for the Integration and Analysis of Multiple High-Throughput "omics" Data.
Chen C, McGarvey PB, Huang H, Wu CH. Chen C, et al. Adv Bioinformatics. 2010;2010:423589. doi: 10.1155/2010/423589. Epub 2010 Mar 29. Adv Bioinformatics. 2010. PMID: 20369061 Free PMC article. - The energetics of genome complexity.
Lane N, Martin W. Lane N, et al. Nature. 2010 Oct 21;467(7318):929-34. doi: 10.1038/nature09486. Nature. 2010. PMID: 20962839 - Exploration of the Germline Genome of the Ciliate Chilodonella uncinata through Single-Cell Omics (Transcriptomics and Genomics).
Maurer-Alcalá XX, Knight R, Katz LA. Maurer-Alcalá XX, et al. mBio. 2018 Jan 9;9(1):e01836-17. doi: 10.1128/mBio.01836-17. mBio. 2018. PMID: 29317511 Free PMC article. - Epigenetics as Driver of Adaptation and Diversification in Microbial Eukaryotes.
Weiner AKM, Katz LA. Weiner AKM, et al. Front Genet. 2021 Mar 16;12:642220. doi: 10.3389/fgene.2021.642220. eCollection 2021. Front Genet. 2021. PMID: 33796133 Free PMC article. No abstract available. - Evolution of germline-limited sequences in two populations of the ciliate Chilodonella uncinata.
Zufall RA, Sturm M, Mahon BC. Zufall RA, et al. J Mol Evol. 2012 Apr;74(3-4):140-6. doi: 10.1007/s00239-012-9493-4. Epub 2012 Mar 13. J Mol Evol. 2012. PMID: 22411695
References
- Adam RD. The Giardia lamblia genome. International Journal for Parasitology. 2000;30:475–484. - PubMed
- Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, Barta JR, Browser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor M. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. Journal of Eukaryotic Microbiology. 2005;52:399–451. - PubMed
- Afonkin SJ. Spontaneous depolyploidization of cells in Amoeba clones with increased nuclear-DNA content. Archiv Für Protistenkunde. 1986;131:101–112.
- Angert ER. Alternatives to binary fission in bacteria. Nature Reviews Microbiology. 2005;3:214–224. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources