The RAST Server: rapid annotations using subsystems technology - PubMed (original) (raw)
doi: 10.1186/1471-2164-9-75.
Daniela Bartels, Aaron A Best, Matthew DeJongh, Terrence Disz, Robert A Edwards, Kevin Formsma, Svetlana Gerdes, Elizabeth M Glass, Michael Kubal, Folker Meyer, Gary J Olsen, Robert Olson, Andrei L Osterman, Ross A Overbeek, Leslie K McNeil, Daniel Paarmann, Tobias Paczian, Bruce Parrello, Gordon D Pusch, Claudia Reich, Rick Stevens, Olga Vassieva, Veronika Vonstein, Andreas Wilke, Olga Zagnitko
Affiliations
- PMID: 18261238
- PMCID: PMC2265698
- DOI: 10.1186/1471-2164-9-75
The RAST Server: rapid annotations using subsystems technology
Ramy K Aziz et al. BMC Genomics. 2008.
Abstract
Background: The number of prokaryotic genome sequences becoming available is growing steadily and is growing faster than our ability to accurately annotate them.
Description: We describe a fully automated service for annotating bacterial and archaeal genomes. The service identifies protein-encoding, rRNA and tRNA genes, assigns functions to the genes, predicts which subsystems are represented in the genome, uses this information to reconstruct the metabolic network and makes the output easily downloadable for the user. In addition, the annotated genome can be browsed in an environment that supports comparative analysis with the annotated genomes maintained in the SEED environment. The service normally makes the annotated genome available within 12-24 hours of submission, but ultimately the quality of such a service will be judged in terms of accuracy, consistency, and completeness of the produced annotations. We summarize our attempts to address these issues and discuss plans for incrementally enhancing the service.
Conclusion: By providing accurate, rapid annotation freely to the community we have created an important community resource. The service has now been utilized by over 120 external users annotating over 350 distinct genomes.
Figures
Figure 1
Example Tricarballylate Utilization Subsystem. A) The subsystem is comprised of 4 functional roles. B) The Subsystem Spreadsheet is populated with genes from 5 organisms (simplified from the original subsystem) where each row represents one organism and each column one functional role. Genes performing the specific functional role in the respective organism populate the respective cell. Gray shading of cells indicates proximity of the respective genes on the chromosomes. There are two distinct variants of the subsystem: variant 1, with all 4 functional roles and variant 2 where the 3rd functional role is missing.
Figure 2
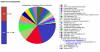
Genes connected to subsystems and their distribution in different categories. The categories are expandable down to the specific gene (see Secondary Metabolism).
Figure 3
Job Overview page. The colours in the progress bar have the following meaning: gray – not started, blue – queued for computation, yellow – in progress, red – requires user input, brown – failed with an error, green – successfully completed.
Figure 4
Job Detail page. The RAST annotation progress can be monitored by each user.
Figure 5
Genome Browser. The annotated genome can be browsed starting from a whole-genome view and zooming-in to a specific feature.
Figure 6
Annotation Overview. For each annotated feature RAST presents an overview page, which includes comparative genomics views and the connections to a subsystem if one was asserted.
Figure 7
Compare Metabolic Reconstruction tool. In the example the RAST metabolic reconstruction of the submitted genome of S. pyogenes Manfredo was compared to the metabolic reconstruction for S. pyogenes MGAS315, which is part of the comparative environment of the SEED. All three columns of subsystem categories are expandable. In cases where RAST was conservative in the assertion of a subsystem a manual attempt to retrieve the missing function/s can be made by clicking the find button.
Figure 8
View Features page. All annotated features can be viewed and downloaded in table format. For each peg the location on the contig, the functional role assignment, its EC number (if present) and GO category, the connection to a subsystem and a KEGG reaction (if appropriate) are given.
Figure 9
View Scenarios page. A genome-specific reaction network can be viewed on a scenario by scenario basis. The scenarios are organized on the left by subsystems, which are themselves organized by categories of metabolic function. If a path through a scenario was found in a given subsystem, the subsystem name is highlighted in blue. In this case, one path was found through the Uroporphyrinogen III generation scenario in the Porphyrin, Heme and Siroheme Biosynthesis subsystem. The table to the right shows the input and output compounds for the scenario, including their stoichiometry, and the reactions that make up the path through the scenario.
Figure 10
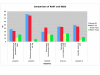
Comparison of a set of genomes manually curated in the SEED and automatically annotated in RAST. The number of genes annotated as hypothetical and the number of genes linked to subsystems (our mechanism of manual curation) is shown to provide an initial assessment of the performance of RAST.
Similar articles
- The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST).
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. Overbeek R, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D206-14. doi: 10.1093/nar/gkt1226. Epub 2013 Nov 29. Nucleic Acids Res. 2014. PMID: 24293654 Free PMC article. - Re-annotation of genome microbial coding-sequences: finding new genes and inaccurately annotated genes.
Bocs S, Danchin A, Médigue C. Bocs S, et al. BMC Bioinformatics. 2002;3:5. doi: 10.1186/1471-2105-3-5. Epub 2002 Feb 5. BMC Bioinformatics. 2002. PMID: 11879526 Free PMC article. - SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models.
Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Stevens RL, Vonstein V, Xia F. Aziz RK, et al. PLoS One. 2012;7(10):e48053. doi: 10.1371/journal.pone.0048053. Epub 2012 Oct 24. PLoS One. 2012. PMID: 23110173 Free PMC article. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review. - Proteogenomics of rare taxonomic phyla: A prospective treasure trove of protein coding genes.
Kumar D, Mondal AK, Kutum R, Dash D. Kumar D, et al. Proteomics. 2016 Jan;16(2):226-40. doi: 10.1002/pmic.201500263. Epub 2015 Nov 23. Proteomics. 2016. PMID: 26773550 Review.
Cited by
- Unveiling a novel exopolysaccharide produced by Pseudomonas alcaligenes Med1 isolated from a Chilean hot spring as biotechnological additive.
Sarkar S, Cabrera-Barjas G, Singh RN, Fabi JP, Breig SJM, Tapia J, Sani RK, Banerjee A. Sarkar S, et al. Sci Rep. 2024 Oct 23;14(1):25058. doi: 10.1038/s41598-024-74830-6. Sci Rep. 2024. PMID: 39443539 Free PMC article. - Characterization of bacteriophage vB_AbaS_SA1 and its synergistic effects with antibiotics against clinical multidrug-resistant Acinetobacter baumannii isolates.
Rastegar S, Sabouri S, Tadjrobehkar O, Samareh A, Niaz H, Sanjari N, Hosseini-Nave H, Skurnik M. Rastegar S, et al. Pathog Dis. 2024 Feb 7;82:ftae028. doi: 10.1093/femspd/ftae028. Pathog Dis. 2024. PMID: 39435653 Free PMC article. - Complete sequence of a bla(KPC-2)-harboring IncFII(K1) plasmid from a Klebsiella pneumoniae sequence type 258 strain.
Chen L, Chavda KD, Melano RG, Jacobs MR, Levi MH, Bonomo RA, Kreiswirth BN. Chen L, et al. Antimicrob Agents Chemother. 2013 Mar;57(3):1542-5. doi: 10.1128/AAC.02332-12. Epub 2013 Jan 7. Antimicrob Agents Chemother. 2013. PMID: 23295924 Free PMC article. - Draft Genome Sequence of Escherichia coli Strain Nissle 1917 (Serovar O6:K5:H1).
Cress BF, Linhardt RJ, Koffas MA. Cress BF, et al. Genome Announc. 2013 Feb 28;1(2):e0004713. doi: 10.1128/genomeA.00047-13. Genome Announc. 2013. PMID: 23516190 Free PMC article. - Description of Chloramphenicol Resistant Kineococcus rubinsiae sp. nov. Isolated From a Spacecraft Assembly Facility.
Mhatre S, Singh NK, Wood JM, Parker CW, Pukall R, Verbarg S, Tindall BJ, Neumann-Schaal M, Venkateswaran K. Mhatre S, et al. Front Microbiol. 2020 Aug 18;11:1957. doi: 10.3389/fmicb.2020.01957. eCollection 2020. Front Microbiol. 2020. PMID: 32973710 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources