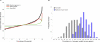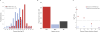Comparing patterns of natural selection across species using selective signatures - PubMed (original) (raw)
Comparative Study
Comparing patterns of natural selection across species using selective signatures
B Jesse Shapiro et al. PLoS Genet. 2008 Feb.
Erratum in
- PLoS Genet. 2008 Jun;4(6). doi: 10.1371/annotation/fced1166-1825-485c-88b9-d6ec950326c8
Abstract
Comparing gene expression profiles over many different conditions has led to insights that were not obvious from single experiments. In the same way, comparing patterns of natural selection across a set of ecologically distinct species may extend what can be learned from individual genome-wide surveys. Toward this end, we show how variation in protein evolutionary rates, after correcting for genome-wide effects such as mutation rate and demographic factors, can be used to estimate the level and types of natural selection acting on genes across different species. We identify unusually rapidly and slowly evolving genes, relative to empirically derived genome-wide and gene family-specific background rates for 744 core protein families in 30 gamma-proteobacterial species. We describe the pattern of fast or slow evolution across species as the "selective signature" of a gene. Selective signatures represent a profile of selection across species that is predictive of gene function: pairs of genes with correlated selective signatures are more likely to share the same cellular function, and genes in the same pathway can evolve in concert. For example, glycolysis and phenylalanine metabolism genes evolve rapidly in Idiomarina loihiensis, mirroring an ecological shift in carbon source from sugars to amino acids. In a broader context, our results suggest that the genomic landscape is organized into functional modules even at the level of natural selection, and thus it may be easier than expected to understand the complex evolutionary pressures on a cell.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
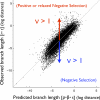
Figure 1. Evolutionary Rate Deviations as Evidence of Natural Selection
Observed branch length is plotted against the branch length predicted from gene-specific (ρ) and species-specific (β) effects (see Methods). A total of 16,681 points are plotted, corresponding to 744 orthologous proteins present in 16–30 species. Amino acid substitutions per site are shown on a log2 scale. The gray line corresponds to y = x.
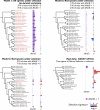
Figure 2. Genes of Common Function Have Similar Selective Signatures
Relative rates of evolution are shown for five genes across 30 species. Fast-evolving genes (log2ν > 0) are shown as red bars; slow-evolving genes (log2ν < 0) as blue bars; genes absent in a given species are not shown. The time scale for the phylogeny was estimated using a Bayesian relaxed molecular clock model [52]. Flagellar genes: flgN (COG 3418; Flagellar biosynthesis/type III secretory pathway chaperone), flgA (COG 1261; Flagellar basal body P-ring biosynthesis protein), fliS (COG 1516; Flagellin-specific chaperone). Sulfur metabolism genes: _yheL (COG 2168; Uncharacterized conserved protein involved in oxidation of intracellular sulfur), yheM (COG 2923; Uncharacterized conserved protein involved in oxidation of intracellular sulfur).
Figure 3. Selection Acts Coherently on Cellular Functions
(A) Correlations in ν, dN/dS, and relative dN/dS (normalized as described in Methods) were obtained for the 109,405 gene-pairs with a COG functional category annotation (16 categories, excluding “general” or “unknown” function). Of these pairs, 10,377 have the same COG function, accounting for a proportion of ∼0.09 of the total (plotted as a solid gray line). Pairs were binned according to correlation-percentile in groups of ten percentile points except for the last three (90%–95%, 95%–99%, 99%–100%). Shown is the fraction with common function in each bin. To avoid potential bias, percentiles were calculated separately for genes present in different numbers of species (15 bins ranging from 16 to 30 species). (B) Gene families under the same model of evolution have highly correlated selective signatures. Correlations in ν were obtained for all pairs of simulated gene families, with or without branch variation in dN/dS, and with dN/dS chosen randomly from within ±1 standard deviation of the mean of the observed dN/dS values (range: 0.005–0.26). The distribution of correlations is shown for pairs of gene families with branch variation in dN/dS and that are replicates of the same evolutionary model (light blue). The distribution of all pairwise correlations—including gene families with or without branch variation, and pairs from the same or different models—is also shown (gray).
Figure 4. Rapidly Evolving Pathways in I. loihiensis
Simplified schematic of glycolysis and phenylalanine metabolism in I. loihiensis. Metabolic intermediates are denoted by white circles; enzymes by arrows. “Fast-evolving” enzymes, depicted as red arrows, are defined as those with ν in the top 10% of genes in the I. loihiensis genome. The names of genes encoding fast-evolving enzymes are shown, highlighted in light blue or orange, respectively for glycolysis or phenylalanine metabolism. Nonfunctional pathways (those with many key enzymes or transporters missing) are shown in gray. Of the “present” enzymes shown in black, only one is slow-evolving (ν < 1) in Idiomarina: COG 191, encoding the enzyme fructose bisphosphate aldolase, which interconverts F1,6P and GA3P. Abbreviations for metabolic intermediates: PEP, phoshphenolpyruvate; E4P, erythrose-4-phosphate; DAHP, 7P-2-dehydro-3-deoxy-arabinoheptonate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; prCat, protocatechuate; shik, shikimate; shik-3P, shikimate-3-phosphate; CVPS, 5-O-(1-carboxyvinyl)-3-phosphoshikimate; chor, chorismate; prePh, prephenate; phPy, phenylpyruvate; Phe, phenylalanine; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6P, fructose-1,6-bisphosphate; GA3P, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetone phosphate; G1,3P, glyerate-1,3-bisphospate; G3P, glycerate-3-phosphate; G2P, glycerate-2-phosphate. COG and EC numbers of fast-evolving genes: AroB: COG337, EC4.2.3.4; AroQ: COG757, EC4.2.1.10; AroE: COG169, EC1.1.1.25; PheA: COG77, EC4.2.1.51; Pgi: COG166, EC5.3.1.9; Fbp: COG158, EC3.1.3.11; Pfk: COG205, EC2.7.1.11; TpiA: COG149, EC5.3.1.1; Eno: COG148, EC4.2.1.11.
Figure 5. Evidence for Positive and Negative Natural Selection
(A) Comparison of relative rates (ν) and Fixation Index. Histograms show the frequency (probability density) distribution of FI values for fast-evolving (ν > 2; dark red; n = 69) and slow-evolving (ν < 0.5; light blue; n = 63) genes. Bins are labelled with the FI value corresponding to their midpoint, on a log2 scale. FI was calculated by counting fixed and polymorphic substitutions at synonymous and nonsynonymous sites, in a sample of 473 COGs (all present in the relatives rates dataset, and consistent with the species tree topology according to the K-H test, as described in the Methods) in 24 _E. coli_ strains, using _S. enterica_ as an outgroup. (B) Purifying selection and gene deletions. Fast-evolvers (or slow-evolvers) were defined as those genes evolving four times faster (or slower) than expected (ν > 4.0 or ν < 0.25, respectively, for fast and slow, with a Z-score > 1.0). For the fast and slow sets of genes, we counted the number with lost orthologs in the closest sister clade in the species tree. When the sister clade contains multiple species, loss indicates the gene was absent from all species in the clade. Frequency of loss among the fast and slow sets was significantly different than the average over all other genes: higher in the fast-evolving set (Fisher's exact test: Odds Ratio = 3.1, p = 2.4e-7), and lower in the slow-evolving set (Fisher's exact test: Odds Ratio = 0.55, p = 0.01). (C) Evidence for genetic hitchhiking. A binomial test was used to determine whether fast (or slow) evolving genes tend to be clustered in the genome near other fast (or slow) evolving genes across all 30 species combined (ν > 1 or ν < 1, respectively, for fast and slow, with a Z-score > 1.0). Log p-values are plotted for pairs separated by distance-windows of 0–5 genes, 6–20 genes, 21–100 genes, 101–200 genes, and 201–300 genes (points shown indicate the maximum separation). The gray line represents p = 0.05.
Figure 6. Detection of Positive Selection by dN/dS and ν under Different Evolutionary Models
Values of dN/dS and ν (mean over 12 replicates of each model) are shown for three simulation models. Model 1: dN/dS = 2 at 3/10 of sites and dN/dS = 1 at 7/10 of sites, in all species (shown in red). Model 2: dN/dS = 2 at 3/10 of sites and dN/dS = 1 at 7/10 of sites, respectively, for the species shown in red. All other branches had dN/dS = 0 at all sites. Model 3: dN/dS = 2, dN/dS = 1, and dN/dS = 0 at 1/10, 7/10, and 2/10 of sites, respectively, in the species shown in red. All other branches had dN/dS = 1 and dN/dS = 0 at 8/10 and 2/10 of sites, respectively. Values of dN/dS and ν are also shown, as estimated for a real protein family from our dataset of 744 protein families in 30 species.
Figure 7. Positive Association of Selective Signatures (ν) and Fixation Index, Independent of dN/dS
We counted E. coli genes with FI > 1.2 or FI < 0.6 as “high” and “low,” and with log2 ν > 0.5 (ν > 1.4) or log2 ν < −0.5 (ν < 0.7) as “high” and “low.” The genes were divided into sets with relatively high dN/dS ( >0.06), medium (0.02 < dN/dS < 0.06), or low dN/dS (<0.02). Within each set, counts were binned in 2 × 2 contingency tables to calculate the Odds Ratio statistic, with Odds Ratio > 1 indicating positive association between ν and FI. One-sided p-values of Fisher's exact test are shown.
Similar articles
- The slow:fast substitution ratio reveals changing patterns of natural selection in gamma-proteobacterial genomes.
Shapiro BJ, Alm E. Shapiro BJ, et al. ISME J. 2009 Oct;3(10):1180-92. doi: 10.1038/ismej.2009.51. Epub 2009 May 21. ISME J. 2009. PMID: 19458656 - Phylogeny of gamma-proteobacteria: resolution of one branch of the universal tree?
Brown JR, Volker C. Brown JR, et al. Bioessays. 2004 May;26(5):463-8. doi: 10.1002/bies.20030. Bioessays. 2004. PMID: 15112225 - Genome sequence of the deep-sea gamma-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy.
Hou S, Saw JH, Lee KS, Freitas TA, Belisle C, Kawarabayasi Y, Donachie SP, Pikina A, Galperin MY, Koonin EV, Makarova KS, Omelchenko MV, Sorokin A, Wolf YI, Li QX, Keum YS, Campbell S, Denery J, Aizawa S, Shibata S, Malahoff A, Alam M. Hou S, et al. Proc Natl Acad Sci U S A. 2004 Dec 28;101(52):18036-41. doi: 10.1073/pnas.0407638102. Epub 2004 Dec 13. Proc Natl Acad Sci U S A. 2004. PMID: 15596722 Free PMC article. - The interplay of homologous recombination and horizontal gene transfer in bacterial speciation.
Lawrence JG, Retchless AC. Lawrence JG, et al. Methods Mol Biol. 2009;532:29-53. doi: 10.1007/978-1-60327-853-9_3. Methods Mol Biol. 2009. PMID: 19271178 Review. - The molecular signature of selection underlying human adaptations.
Harris EE, Meyer D. Harris EE, et al. Am J Phys Anthropol. 2006;Suppl 43:89-130. doi: 10.1002/ajpa.20518. Am J Phys Anthropol. 2006. PMID: 17103426 Review.
Cited by
- In silico functional and evolutionary analyses of rubber oxygenases (RoxA and RoxB).
Sharma V, Mobeen F, Prakash T. Sharma V, et al. 3 Biotech. 2020 Sep;10(9):376. doi: 10.1007/s13205-020-02371-6. Epub 2020 Aug 5. 3 Biotech. 2020. PMID: 32802718 Free PMC article. - Assessment of Genetic Diversity and Conservation in South African Indigenous Goat Ecotypes: A Review.
Magoro AM, Mtileni B, Hadebe K, Zwane A. Magoro AM, et al. Animals (Basel). 2022 Nov 29;12(23):3353. doi: 10.3390/ani12233353. Animals (Basel). 2022. PMID: 36496874 Free PMC article. Review. - Diversity Takes Shape: Understanding the Mechanistic and Adaptive Basis of Bacterial Morphology.
Kysela DT, Randich AM, Caccamo PD, Brun YV. Kysela DT, et al. PLoS Biol. 2016 Oct 3;14(10):e1002565. doi: 10.1371/journal.pbio.1002565. eCollection 2016 Oct. PLoS Biol. 2016. PMID: 27695035 Free PMC article. - Covariation of branch lengths in phylogenies of functionally related genes.
Li WL, Rodrigo AG. Li WL, et al. PLoS One. 2009 Dec 29;4(12):e8487. doi: 10.1371/journal.pone.0008487. PLoS One. 2009. PMID: 20041191 Free PMC article. - Tracking Microbial Evolution in the Subseafloor Biosphere.
Anderson RE. Anderson RE. mSystems. 2021 Aug 31;6(4):e0073121. doi: 10.1128/mSystems.00731-21. Epub 2021 Aug 17. mSystems. 2021. PMID: 34402637 Free PMC article.
References
- Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, et al. A genome-wide map of diversity in plasmodium falciparum. Nat Genet. 2007;39:113–119. - PubMed
- Thornton KR, Jensen JD, Becquet C, Andolfatto P. Progress and prospects in mapping recent selection in the genome. Heredity. 2007;98:340–348. - PubMed