Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana - PubMed (original) (raw)
Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana
Carey F Li et al. PLoS Genet. 2008 Feb.
Abstract
DNA methylation directed by 24-nucleotide small RNAs involves the small RNA-binding protein ARGONAUTE4 (AGO4), and it was previously shown that AGO4 localizes to nucleolus-adjacent Cajal bodies, sites of snRNP complex maturation. Here we demonstrate that AGO4 also localizes to a second class of nuclear bodies, called AB-bodies, which are found immediately adjacent to condensed 45S ribosomal DNA (rDNA) sequences. AB-bodies also contain other proteins involved in RNA-directed DNA methylation including NRPD1b (a subunit of the RNA Polymerase IV complex, RNA PolIV), NRPD2 (a second subunit of this complex), and the DNA methyltransferase DRM2. These two classes of AGO4 bodies are structurally independent--disruption of one class does not affect the other--suggesting a dynamic regulation of AGO4 within two distinct nuclear compartments in Arabidopsis. Abolishing Cajal body formation in a coilin mutant reduced overall AGO4 protein levels, and coilin dicer-like3 double mutants showed a small decrease in DNA methylation beyond that seen in dicer-like3 single mutants, suggesting that Cajal bodies are required for a fully functioning DNA methylation system in Arabidopsis.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
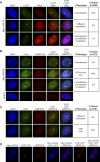
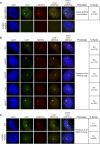
Figure 1. AGO4 Localizes to Two Distinct Nuclear Bodies
(A) Fluorescent microscopy analysis of Myc-AGO4 and Cajal body localization in nuclei isolated from Myc-AGO4 seedlings. A monoclonal antibody to endogenous U2B′′ was used to detect the Cajal body. Three different AGO4 localization patterns relative to U2B′′ are shown. Nuclei containing observable AGO4 nuclear foci were examined. (B) Localization of Myc-AGO4 and NRPD1b in Myc-AGO4 nuclei. A polyclonal antibody was used to detect endogenous NRPD1b. Three representative nuclei are shown. Nuclei containing observable AGO4 nuclear foci were examined. (C) Localization of U2B′′ relative to NRPD1b within wild type Ler nuclei. (D) Three color fluorescent microscopy analysis of Myc-AGO4, U2B′′-GFP, and NRPD1b localization within the same nucleus. One representative nucleus containing two different AGO4 bodies is shown: colocalization with NRPD1b (top of the nucleus) versus colocalization with the Cajal body (bottom of the nucleus). All AGO4 foci colocalized with either NRPD1b or Cajal body (n = 156).
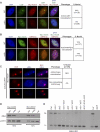
Figure 2. Effects of the Cajal Body on AGO4 Localization and DNA Methylation
(A,B) Immunofluorescence analysis of Myc-AGO4 localization in wild type nuclei or a coilin mutant (SALK 148589) is shown relative to (A) U2B′′ or (B) NRPD1b. (C) Myc-AGO4 nuclear distribution in a plant line overexpressing COILIN-mRFP and containing enlarged Cajal bodies. (D) Semi-quantitative western blot analysis of Myc-AGO4 protein. Total protein amounts of 17 μg (labeled as 100%) or 8.5 μg (labeled as 50%) were loaded per sample. An antibody to c-Myc (Upstate) was used to detect Myc-AGO4. The endogenous photoreceptor CRY1 was used as a loading control [47]. (E) Southern blot analysis examining DNA methylation at the MEA-ISR locus. Genomic DNA was digested with methyl-sensitive enzyme _Msp_I (mCCGG). Three independent biological replicates of dcl3 and ncb-1 dcl3 were examined. The band indicating methylated DNA (undigested) is labeled as “M”, while the band indicating unmethylated DNA (digested) is labeled as “U”.
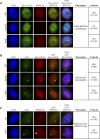
Figure 3. Presence of the AB-Body in Upstream RdDM Mutants
Immunolocalization analysis of Myc-AGO4 and NRPD1b in wild type, dcl3, rdr2, dcl3 rdr2, or nrpd1a nuclei. White arrows indicate the presence of a faintly staining Myc-AGO4 body that colocalized with NRPD1b. Two populations of nuclei are shown for each mutant: nuclei with and nuclei without AB-bodies.
Figure 4. Effects of Downstream RdDM Mutations on the AB-Body
(A,B) Myc-AGO4 nuclear localization in wild type, nrpd1b, or drm2 mutant nuclei is shown relative to the immunostaining of (A) NRPD1b or (B) U2B′′. (C) Immunolocalization of NRPD1b and U2B′′ in wild type (Ler) or an ago4 mutant. The white arrow indicates a less intense NRPD1b body that still remained in ago4.
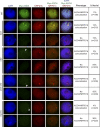
Figure 5. Localization of NRPD2 Relative to the AB-Body
(A) Immunofluorescence analysis of NRPD2 localization relative to AGO4 in Myc-AGO4 nuclei. A polyclonal antibody was used to detect endogenous NRPD2 protein. (B) Localization of NRPD2 relative to U2B′′ in wild type, nrpd1a, nrpd1b, or ago4 mutant nuclei. Ler is the wild type control for ago4 within the same ecotype, and Col is the wild type control for nrpd1a and nrpd1b. (C) NRPD1b nuclear body formation in wild type versus nrpd2a nrpd2b. Nuclei were co-immunostained with an antibody to U2B′′.
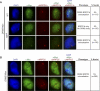
Figure 6. Localization of AB-Bodies to the Immediate Proximity of the 45S rDNA Loci
(A) DNA FISH analysis of the condensed 45S rDNA loci (NORs) relative to Myc-AGO4 (left) or NRPD1b (right) localization in Myc-AGO4 or Ler nuclei, respectively. (B,C) Localization of AGO4 or NRPD1b is shown relative to (B) 5S rDNA loci or (C) centromeric repeats (CEN). (D) Dual probe DNA FISH analysis showing 45S and 5S rDNA loci relative to Myc-AGO4 (top panel) or NRPD1b (bottom panel). The 45S probe was labeled with biotin and the 5S probe was labeled with digoxigenin (DIG). NOR4 is observed as a 45S rDNA signal with an adjacent 5S rDNA signal, while NOR2 shows only 45S rDNA signal with no adjacent 5S rDNA signal.
Figure 7. Localization of DRM2 with AB-odies
(A) DRM2-Myc nuclear localization relative to NRPD1b in wild type or an ago4 mutant. (B) Immunofluorescence analysis of the NRPD2 nuclear body relative to the DRM2 body.
Similar articles
- An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana.
Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. Li CF, et al. Cell. 2006 Jul 14;126(1):93-106. doi: 10.1016/j.cell.2006.05.032. Cell. 2006. PMID: 16839879 - The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center.
Pontes O, Li CF, Costa Nunes P, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. Pontes O, et al. Cell. 2006 Jul 14;126(1):79-92. doi: 10.1016/j.cell.2006.05.031. Cell. 2006. PMID: 16839878 - Cell biology of the Arabidopsis nuclear siRNA pathway for RNA-directed chromatin modification.
Pikaard CS. Pikaard CS. Cold Spring Harb Symp Quant Biol. 2006;71:473-80. doi: 10.1101/sqb.2006.71.046. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381329 Review. - NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation.
He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. He XJ, et al. Genes Dev. 2009 Feb 1;23(3):318-30. doi: 10.1101/gad.1765209. Genes Dev. 2009. PMID: 19204117 Free PMC article. - RNA-directed DNA methylation and Pol IVb in Arabidopsis.
Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Matzke M, et al. Cold Spring Harb Symp Quant Biol. 2006;71:449-59. doi: 10.1101/sqb.2006.71.028. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381327 Review.
Cited by
- Biomolecular condensates in plant RNA silencing: insights into formation, function, and stress responses.
Li Q, Liu Y, Zhang X. Li Q, et al. Plant Cell. 2024 Jan 30;36(2):227-245. doi: 10.1093/plcell/koad254. Plant Cell. 2024. PMID: 37772963 Free PMC article. Review. - Molecular cloning and characterisation of SlAGO family in tomato.
Xian Z, Yang Y, Huang W, Tang N, Wang X, Li Z. Xian Z, et al. BMC Plant Biol. 2013 Sep 8;13:126. doi: 10.1186/1471-2229-13-126. BMC Plant Biol. 2013. PMID: 24011258 Free PMC article. - Connecting the dots of RNA-directed DNA methylation in Arabidopsis thaliana.
Costa-Nunes P, Vitins A, Pontes O. Costa-Nunes P, et al. Chromosome Res. 2014 Jun;22(2):225-40. doi: 10.1007/s10577-014-9425-9. Chromosome Res. 2014. PMID: 24846724 Review. - How intrinsically disordered proteins order plant gene silencing.
Shang B, Li C, Zhang X. Shang B, et al. Trends Genet. 2024 Mar;40(3):260-275. doi: 10.1016/j.tig.2023.12.009. Epub 2024 Jan 30. Trends Genet. 2024. PMID: 38296708 Review. - Role of small RNAs in host-microbe interactions.
Katiyar-Agarwal S, Jin H. Katiyar-Agarwal S, et al. Annu Rev Phytopathol. 2010;48:225-46. doi: 10.1146/annurev-phyto-073009-114457. Annu Rev Phytopathol. 2010. PMID: 20687832 Free PMC article. Review.
References
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. - PubMed
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. - PubMed
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. - PubMed
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM060398/GM/NIGMS NIH HHS/United States
- GM07185/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 GM007185/GM/NIGMS NIH HHS/United States
- R01 GM060398/GM/NIGMS NIH HHS/United States
- GM60398/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases