Genetic and chemical modifiers of a CUG toxicity model in Drosophila - PubMed (original) (raw)
Genetic and chemical modifiers of a CUG toxicity model in Drosophila
Amparo Garcia-Lopez et al. PLoS One. 2008.
Abstract
Non-coding CUG repeat expansions interfere with the activity of human Muscleblind-like (MBNL) proteins contributing to myotonic dystrophy 1 (DM1). To understand this toxic RNA gain-of-function mechanism we developed a Drosophila model expressing 60 pure and 480 interrupted CUG repeats in the context of a non-translatable RNA. These flies reproduced aspects of the DM1 pathology, most notably nuclear accumulation of CUG transcripts, muscle degeneration, splicing misregulation, and diminished Muscleblind function in vivo. Reduced Muscleblind activity was evident from the sensitivity of CUG-induced phenotypes to a decrease in muscleblind genetic dosage and rescue by MBNL1 expression, and further supported by the co-localization of Muscleblind and CUG repeat RNA in ribonuclear foci. Targeted expression of CUG repeats to the developing eye and brain mushroom bodies was toxic leading to rough eyes and semilethality, respectively. These phenotypes were utilized to identify genetic and chemical modifiers of the CUG-induced toxicity. 15 genetic modifiers of the rough eye phenotype were isolated. These genes identify putative cellular processes unknown to be altered by CUG repeat RNA, and they include mRNA export factor Aly, apoptosis inhibitor Thread, chromatin remodelling factor Nurf-38, and extracellular matrix structural component Viking. Ten chemical compounds suppressed the semilethal phenotype. These compounds significantly improved viability of CUG expressing flies and included non-steroidal anti-inflammatory agents (ketoprofen), muscarinic, cholinergic and histamine receptor inhibitors (orphenadrine), and drugs that can affect sodium and calcium metabolism such as clenbuterol and spironolactone. These findings provide new insights into the DM1 phenotype, and suggest novel candidates for DM1 treatments.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. Flies expressing CUG repeats show shorter lifespan.
Average percentage of live flies, with the genotypes indicated, versus age (in days). (A) Whereas control flies showed an average lifespan of 57 (UAS-i(CTG)480/+; n = 80) and 34 (Mhc-Gal4/+; n = 40) days, i(CUG)480-expressing flies lived 13 days in average (n = 60). Differences in lifespan curves were highly significant when comparing i(CUG)480-expressing flies to UAS-i(CTG)480/+ control flies (p<0.0001, Log-Rank test) but not to Mhc-Gal4/+_ controls. (B) Expression of i(CUG)480 transgene in an ubiquitous manner (_da-Gal4>UAS-i(CTG)480) also reduced fly survival. Control flies showed a median survival of 57 (UAS-i(CTG)480/+, n = 80) and 55 (da-Gal4/+, n = 40) days. Median survival for i(CUG)480-expressing flies was of 41 days (n = 80), and lifespan curves for i(CUG)480-expressing flies and both controls showed differences that were statistically significant (p<0.0001, Log-Rank test). Statistical analysis was performed using GraphPad Prism4 software.
Figure 2. CUG-induced eye and muscle degeneration in flies.
Transversal sections of resin-embedded (A–F) adult IFMs of control flies (Mhc-Gal4/+) (A, D) and flies expressing (CUG)60 (B, E) or i(CUG)480 RNA (C, F) under the control of the Mhc-Gal4 driver. IFMs were studied in 2–3-day old (A–C) or 38-day old (D–F) flies. Expression of (CUG)60 RNA was not toxic to muscle fibres (B), and IFMs did not degenerate over time (E). Expression of i(CUG)480 RNA in IFMs led to vacuolization (arrowheads) and muscle disorganization (C). Muscle degeneration and wasting was conspicuous in 38-day old flies with large vacuoles (arrowheads), lower density of myofibrils per muscle (arrow) and missing muscles (asterisk). Results consistent with these have been obtained independently . (G) Cross-sectional area of left dorso longitudinal muscle 45e in 2 day-old control (Mhc-Gal4/+) and DM1 model flies (Mhc-Gal4>UAS-i(CTG)480). n = 12 (control) and n = 34 (CUG expressing). (H) Muscle degeneration was measured as the frequency of vacuolar pathology and muscle area reduction, according to the following rating scale: vacuoles with diameter larger or smaller than 8 µm, or showing 45% or less of the normal muscle area. Muscle 45e was measured in 3-4 thorax sections per animal and a total of 15 young (2-day-old) or 13 aged (38-day-old) flies were analyzed. Control (Mhc-Gal4/+) and Mhc-Gal4>UAS-(CTG)60 flies showed no muscle phenotype 2 or 38 days after eclosion. Tangential (I, J) and frontal (K, L) sections of adult Drosophila eyes with the genotypes gmr-Gal4/+ (I, K) and gmr-Gal4/UAS-i(CUG)480 (J, L) at 25°C. (I) Tangential sections exhibited a normal complement of photoreceptors per ommatidial unit (arrowheads point to rhabdomeres), although some pigment cells were absent. (J) Expression of expanded CUG repeats caused general disorganization of the eye retina. (K) In controls, rhabdomeres extend from the apical to the basal side of the retina (arrowheads) and the layer of pigment cell feet forms the fenestrated membrane (arrow), which is separated by the basement membrane (white arrowhead) from the underlying subretinal cells (bent arrow). (L) Eyes expressing i(CUG)480 RNA lacked rhabdomeres and showed general disorganization of pigment cells. Fenestrated membranes were thinner and showed gaps (arrow). Subretinal cells were not tightly apposed to the basement membrane (bent arrow).
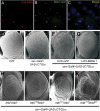
Figure 3. Muscleblind forms nuclear inclusions with CUG repeat RNA and genetically interacts with repeat RNA phenotypes in vivo.
i(CUG)480 RNA and MblC:GFP were coexpressed in adult flies using a hs-Gal4 line (A–C). i(CUG)480 transcripts detected by FISH (red; A) and MblC:GFP detected by the GFP tag (green; B). Red and green channels are shown merged in (C), with nuclei counterstained with DAPI. Scanning electron microscope (SEM) images of Drosophila eyes (D–K). (D) External morphology of reference strain OrR. (E) _sev_-Gal4 driven expression of (CUG)60 RNA in eye precursors exhibits mild external defects, only altering mechanosensory bristles. Expression of i(CUG)480 RNA driven by the same Gal4 generates a rough and reduced eye (H), a phenotype that is specifically suppressed by the simultaneous expression of human MBNL1 (G) but not by expression of the unrelated GFP protein (F). The CUG-dependent eye phenotype was not modified by the weak mbl7103 allele (I), and only slightly modified by mblE27 (J). However, the compound heterozygote enhanced roughness and eye size reduction (K). The compound heterozygote mbl7103/mblE27 displayed normal eyes in the absence of i(CUG)480 RNA.
Figure 4. CUG repeat RNA misregulates alternative splicing of muscle genes CG30084 and TnT.
RT-PCR products from CG30084 (A) and TnT (B) at the stages and from animals with the genotypes indicated. Bar graph representing intensities of ethidium bromide fluorescence (ranging from 0 to 100%, which equalled saturation) of band E (CG30084; C) and band D (TnT; D) from the specified genotypes. All RT-PCRs were within the linear range of amplification. Abbreviations used: 16–18 h after egg laying embryos (E); 2-day old pupae (P); 6–30 h after eclosion adults (A). All missplicing events were detected at least twice from independent RNA extractions.
Figure 5. Dominant genetic enhancers and chemical suppressors of CUG-induced phenotypes.
Stereomicroscope (A, B) and SEM (C–H) views of adult Drosophila eyes. Female flies with the genotype sev-Gal4 UAS-i(CTG)480/+ (A, C, E) show eyes smaller than normal and externally rough. Both features increased in female flies heterozygous for svp1 (B), Aly02267 (D), thread4 (F) and vikingk00236 (H) in the same genetic background whereas overexpression of th (thEP3308, G) considerably improved morphology. Flies were raised at 25°C. (I) Percentage of viable females from crosses between the X-linked 103Y-Gal4 line and lines carrying the UAS-i(CTG)480 or UAS-(CTG)60 transgenes (note that only F1 females express CUG RNA) at different temperatures. Only expression of 480 CUG RNA exhibited a temperature-dependent semilethal phenotype. Three independent MB-specific Gal4 driver lines showed a similar behaviour. (J) Emerged/non-emerged ratio measures the likelihood of survival of CUG-expressing females in control (−drug) and drug-treated flies (+drug). Abbreviations: 1, spironolactone; 2, metoclopramide; 3, ketoprofen; 4, nefopam; 5, orphenadrine; 6, proglumide; 7, ethisterone; 8, indomethacin; 9, clenbuterol; 10, thioguanosine.
Similar articles
- MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1.
de Haro M, Al-Ramahi I, De Gouyon B, Ukani L, Rosa A, Faustino NA, Ashizawa T, Cooper TA, Botas J. de Haro M, et al. Hum Mol Genet. 2006 Jul 1;15(13):2138-45. doi: 10.1093/hmg/ddl137. Epub 2006 May 24. Hum Mol Genet. 2006. PMID: 16723374 - Muscleblind, BSF and TBPH are mislocalized in the muscle sarcomere of a Drosophila myotonic dystrophy model.
Llamusi B, Bargiela A, Fernandez-Costa JM, Garcia-Lopez A, Klima R, Feiguin F, Artero R. Llamusi B, et al. Dis Model Mech. 2013 Jan;6(1):184-96. doi: 10.1242/dmm.009563. Epub 2012 Nov 1. Dis Model Mech. 2013. PMID: 23118342 Free PMC article. - Muscleblind isoforms are functionally distinct and regulate alpha-actinin splicing.
Vicente M, Monferrer L, Poulos MG, Houseley J, Monckton DG, O'dell KM, Swanson MS, Artero RD. Vicente M, et al. Differentiation. 2007 Jun;75(5):427-40. doi: 10.1111/j.1432-0436.2006.00156.x. Epub 2007 Feb 16. Differentiation. 2007. PMID: 17309604 - The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing.
Pascual M, Vicente M, Monferrer L, Artero R. Pascual M, et al. Differentiation. 2006 Mar;74(2-3):65-80. doi: 10.1111/j.1432-0436.2006.00060.x. Differentiation. 2006. PMID: 16533306 Review. - Short Tandem Repeat Expansions and RNA-Mediated Pathogenesis in Myotonic Dystrophy.
Sznajder ŁJ, Swanson MS. Sznajder ŁJ, et al. Int J Mol Sci. 2019 Jul 9;20(13):3365. doi: 10.3390/ijms20133365. Int J Mol Sci. 2019. PMID: 31323950 Free PMC article. Review.
Cited by
- Methylphenidate Attenuates the Cognitive and Mood Alterations Observed in Mbnl2 Knockout Mice and Reduces Microglia Overexpression.
Ramon-Duaso C, Gener T, Consegal M, Fernández-Avilés C, Gallego JJ, Castarlenas L, Swanson MS, de la Torre R, Maldonado R, Puig MV, Robledo P. Ramon-Duaso C, et al. Cereb Cortex. 2019 Jul 5;29(7):2978-2997. doi: 10.1093/cercor/bhy164. Cereb Cortex. 2019. PMID: 30060068 Free PMC article. - Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target.
Krzyzosiak WJ, Sobczak K, Wojciechowska M, Fiszer A, Mykowska A, Kozlowski P. Krzyzosiak WJ, et al. Nucleic Acids Res. 2012 Jan;40(1):11-26. doi: 10.1093/nar/gkr729. Epub 2011 Sep 9. Nucleic Acids Res. 2012. PMID: 21908410 Free PMC article. Review. - Deregulations of miR-1 and its target Multiplexin promote dilated cardiomyopathy associated with myotonic dystrophy type 1.
Souidi A, Nakamori M, Zmojdzian M, Jagla T, Renaud Y, Jagla K. Souidi A, et al. EMBO Rep. 2023 Apr 5;24(4):e56616. doi: 10.15252/embr.202256616. Epub 2023 Feb 28. EMBO Rep. 2023. PMID: 36852954 Free PMC article. - Dissecting Pathogenetic Mechanisms and Therapeutic Strategies in Drosophila Models of Myotonic Dystrophy Type 1.
Souidi A, Zmojdzian M, Jagla K. Souidi A, et al. Int J Mol Sci. 2018 Dec 18;19(12):4104. doi: 10.3390/ijms19124104. Int J Mol Sci. 2018. PMID: 30567354 Free PMC article. Review. - Silencing of drpr leads to muscle and brain degeneration in adult Drosophila.
Draper I, Mahoney LJ, Mitsuhashi S, Pacak CA, Salomon RN, Kang PB. Draper I, et al. Am J Pathol. 2014 Oct;184(10):2653-61. doi: 10.1016/j.ajpath.2014.06.018. Epub 2014 Aug 8. Am J Pathol. 2014. PMID: 25111228 Free PMC article.
References
- Harper P. London: Saunders; 2001. Myotonic dystrophy.
- Ranum LP, Cooper TA. RNA-Mediated Neuromuscular Disorders. Annu Rev Neurosci 2006 - PubMed
- Mankodi A, Logigian E, Callahan L, McClain C, White R, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. - PubMed
- Houseley JM, Wang Z, Brock GJ, Soloway J, Artero R, et al. Myotonic dystrophy associated expanded CUG repeat muscleblind positive ribonuclear foci are not toxic to Drosophila. Hum Mol Genet. 2005;14:873–883. - PubMed
- Haro MD, Al-Ramahi I, Gouyon BD, Ukani L, Rosa A, et al. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of Myotonic Dystrophy Type 1. Hum Mol Genet 2006 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases