A three-stage colonization model for the peopling of the Americas - PubMed (original) (raw)
A three-stage colonization model for the peopling of the Americas
Andrew Kitchen et al. PLoS One. 2008.
Abstract
Background: We evaluate the process by which the Americas were originally colonized and propose a three-stage model that integrates current genetic, archaeological, geological, and paleoecological data. Specifically, we analyze mitochondrial and nuclear genetic data by using complementary coalescent models of demographic history and incorporating non-genetic data to enhance the anthropological relevance of the analysis.
Methodology/findings: Bayesian skyline plots, which provide dynamic representations of population size changes over time, indicate that Amerinds went through two stages of growth approximately 40,000 and approximately 15,000 years ago separated by a long period of population stability. Isolation-with-migration coalescent analyses, which utilize data from sister populations to estimate a divergence date and founder population sizes, suggest an Amerind population expansion starting approximately 15,000 years ago.
Conclusions/significance: These results support a model for the peopling of the New World in which Amerind ancestors diverged from the Asian gene pool prior to 40,000 years ago and experienced a gradual population expansion as they moved into Beringia. After a long period of little change in population size in greater Beringia, Amerinds rapidly expanded into the Americas approximately 15,000 years ago either through an interior ice-free corridor or along the coast. This rapid colonization of the New World was achieved by a founder group with an effective population size of approximately 1,000-5,400 individuals. Our model presents a detailed scenario for the timing and scale of the initial migration to the Americas, substantially refines the estimate of New World founders, and provides a unified theory for testing with future datasets and analytic methods.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
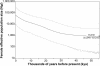
Figure 1. Bayesian skyline plot for the mtDNA coding genome sequences.
The curve plots median Nef with its 95% CI indicated by the light gray lines. The calculated Nef assumes a generation time of 20 years following Hey ; alternatively, using a generation time of 25 years would uniformly decrease all estimates of Nef by 20%. “X” marks the median coalescent time with its 95% CI given in brackets. The shaded regions highlight two periods of substantial population growth. This skyline plot provides the principal evidence for our three-stage model of New World colonization, i.e. the three stages that are depicted and labeled here.
Figure 2. Bayesian skyline plot for the mtDNA HVR I+II datasets.
This plot follows the conventions of Figure 1. Its estimates of coalescent time and Nef at the coalescence and today are in agreement with the coding mtDNA skyline plot (Figure 1). In contrast, this HVRI+II plot provides little resolution for other population size changes, most likely because of mutational saturation in the non-coding control region (see text).
Figure 3. Graph of IM results for the combined nuclear and mitochondrial coding DNA dataset.
The plot depicts mean Ne for the Amerind founder population (y-axis) as a product of increasing the constraint on the upper bound of the priors for the migration rates (x-axis). In these analyses, the prior on the lower bound of the divergence time was uniformly set to 15 kya on the basis of known archaeological materials for human occupation in the New World (see text). Each point is based on the average of the estimated medians for ten independent replicate analyses, with the bars corresponding to ± 1 standard deviation. These standard deviations are often small (with coefficients of variation less than 0.01), since their Markov chains were run for 100 million generations each.
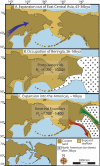
Figure 4. Maps depicting each phase of our three-step colonization model for the peopling of the Americas.
(A) Divergence, then gradual population expansion of the Amerind ancestors from their East Central Asian gene pool (blue arrow). (B) Proto-Amerind occupation of Beringia with little to no population growth for ≈20,000 years. (C) Rapid colonization of the New World by a founder group migrating southward through the ice free, inland corridor between the eastern Laurentide and western Cordilleran Ice Sheets (green arrow) and/or along the Pacific coast (red arrow). In (B), the exposed seafloor is shown at its greatest extent during the last glacial maximum at ≈20–18 kya . In (A) and (C), the exposed seafloor is depicted at ≈40 kya and ≈16 kya, when prehistoric sea levels were comparable , . Because of the earth's curvature, the km scale (which is based on the straight line distance at the equator) provides only an approximation of the same distance between two points on these maps. In addition, a scaled-down version of Beringia today (60% reduction of A–C) is presented in the lower left corner. This smaller map highlights the Bering Strait that has geographically separated the New World from Asia since ≈11–10 kya.
Similar articles
- Updated three-stage model for the peopling of the Americas.
Mulligan CJ, Kitchen A, Miyamoto MM. Mulligan CJ, et al. PLoS One. 2008 Sep 17;3(9):e3199. doi: 10.1371/journal.pone.0003199. PLoS One. 2008. PMID: 18797500 Free PMC article. - Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas.
Llamas B, Fehren-Schmitz L, Valverde G, Soubrier J, Mallick S, Rohland N, Nordenfelt S, Valdiosera C, Richards SM, Rohrlach A, Romero MI, Espinoza IF, Cagigao ET, Jiménez LW, Makowski K, Reyna IS, Lory JM, Torrez JA, Rivera MA, Burger RL, Ceruti MC, Reinhard J, Wells RS, Politis G, Santoro CM, Standen VG, Smith C, Reich D, Ho SY, Cooper A, Haak W. Llamas B, et al. Sci Adv. 2016 Apr 1;2(4):e1501385. doi: 10.1126/sciadv.1501385. eCollection 2016 Apr. Sci Adv. 2016. PMID: 27051878 Free PMC article. - Current evidence allows multiple models for the peopling of the Americas.
Potter BA, Baichtal JF, Beaudoin AB, Fehren-Schmitz L, Haynes CV, Holliday VT, Holmes CE, Ives JW, Kelly RL, Llamas B, Malhi RS, Miller DS, Reich D, Reuther JD, Schiffels S, Surovell TA. Potter BA, et al. Sci Adv. 2018 Aug 8;4(8):eaat5473. doi: 10.1126/sciadv.aat5473. eCollection 2018 Aug. Sci Adv. 2018. PMID: 30101195 Free PMC article. Review. - A single and early migration for the peopling of the Americas supported by mitochondrial DNA sequence data.
Bonatto SL, Salzano FM. Bonatto SL, et al. Proc Natl Acad Sci U S A. 1997 Mar 4;94(5):1866-71. doi: 10.1073/pnas.94.5.1866. Proc Natl Acad Sci U S A. 1997. PMID: 9050871 Free PMC article. - The late Pleistocene dispersal of modern humans in the Americas.
Goebel T, Waters MR, O'Rourke DH. Goebel T, et al. Science. 2008 Mar 14;319(5869):1497-502. doi: 10.1126/science.1153569. Science. 2008. PMID: 18339930 Review.
Cited by
- The social lives of isolates (and small language families): the case of the Northwest Amazon.
Van Gijn R, Norder S, Arias L, Emlen NQ, Azevedo MCBC, Caine A, Dunn S, Howard A, Julmi N, Krasnoukhova O, Stoneking M, Wiegertjes J. Van Gijn R, et al. Interface Focus. 2022 Dec 9;13(1):20220054. doi: 10.1098/rsfs.2022.0054. eCollection 2023 Feb 6. Interface Focus. 2022. PMID: 36655194 Free PMC article. - A genomic perspective on South American human history.
Silva MACE, Ferraz T, Hünemeier T. Silva MACE, et al. Genet Mol Biol. 2022 Jul 29;45(3 Suppl 1):e20220078. doi: 10.1590/1678-4685-GMB-2022-0078. eCollection 2022. Genet Mol Biol. 2022. PMID: 35925590 Free PMC article. - Novel alleles gained during the Beringian isolation period.
Niedbalski SD, Long JC. Niedbalski SD, et al. Sci Rep. 2022 Mar 11;12(1):4289. doi: 10.1038/s41598-022-08212-1. Sci Rep. 2022. PMID: 35277570 Free PMC article. - Peopling of the Americas as inferred from ancient genomics.
Willerslev E, Meltzer DJ. Willerslev E, et al. Nature. 2021 Jun;594(7863):356-364. doi: 10.1038/s41586-021-03499-y. Epub 2021 Jun 16. Nature. 2021. PMID: 34135521 Review. - Moose genomes reveal past glacial demography and the origin of modern lineages.
Dussex N, Alberti F, Heino MT, Olsen RA, van der Valk T, Ryman N, Laikre L, Ahlgren H, Askeyev IV, Askeyev OV, Shaymuratova DN, Askeyev AO, Döppes D, Friedrich R, Lindauer S, Rosendahl W, Aspi J, Hofreiter M, Lidén K, Dalén L, Díez-Del-Molino D. Dussex N, et al. BMC Genomics. 2020 Dec 2;21(1):854. doi: 10.1186/s12864-020-07208-3. BMC Genomics. 2020. PMID: 33267779 Free PMC article.
References
- Greenberg JH, Turner CG, Zegura SL. The settlement of the Americas: a comparison of the linguistic, dental and genetic evidence. Curr Anthropol. 1986;27:477–497.
- Schurr TG. The peopling of the New World: perspectives from molecular anthropology. Annu Rev Anthropol. 2004;33:551–583.
- Mulligan CJ, Hunley K, Cole S, Long JC. Population genetics, history, and health patterns in native Americans. Annu Rev Genom Hum Genet. 2004;5:295–315. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources