Close correspondence between the motions from principal component analysis of multiple HIV-1 protease structures and elastic network modes - PubMed (original) (raw)
Close correspondence between the motions from principal component analysis of multiple HIV-1 protease structures and elastic network modes
Lei Yang et al. Structure. 2008 Feb.
Abstract
The large number of available HIV-1 protease structures provides a remarkable sampling of conformations of the different conformational states, which can be viewed as direct structural information about the dynamics of the HIV-1 protease. After structure matching, we apply principal component analysis (PCA) to obtain the important apparent motions for both bound and unbound structures. There are significant similarities between the first few key motions and the first few low-frequency normal modes calculated from a static representative structure with an elastic network model (ENM), strongly suggesting that the variations among the observed structures and the corresponding conformational changes are facilitated by the low-frequency, global motions intrinsic to the structure. Similarities are also found when the approach is applied to an NMR ensemble, as well as to molecular dynamics (MD) trajectories. Thus, a sufficiently large number of experimental structures can directly provide important information about protein dynamics, but ENM can also provide similar sampling of conformations.
Figures
Figure 1
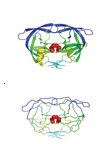
Cartoon representation (A) and alpha carbon trace (B) of the HIV-1 protease structure. Blue - the flap domain; green - the core domain; cyan - the terminal domain; yellow - other residues. The red spheres represent the conserved Asp25-Thr26-Gly27 active site triad. The figure was created using PyMOL (DeLano Scientific).
Figure 2
RMSD with respect to the reference structure for: (A) X-ray-I dataset, with the RMSD values sorted in ascending order. X-ray-II dataset is the same as X-ray-I, excluding the eight structures that have significantly larger RMSD values than the rest. (B) NMR dataset, sorted by the RMSD values in ascending order. (C) MD dataset, shown in the order of the time steps along the 10 ns simulation. (D) MD dataset, sorted by the RMSD values in ascending order.
Figure 3
The fraction of variance (‘o’) and the cumulative fraction of variance (‘x’) represented by the first 6 PCs for: (A) X-ray-I dataset. (B) X-ray-II dataset. (C) NMR dataset. (D) MD dataset.
Figure 4
Distribution of individual structures along pairs of the first three principal component directions. Shown are the planes of PC 1 and PC 2 and of PC 1 and PC 3 for X-ray-I, X-ray-II, NMR and MD datasets respectively. (For the MD dataset, the 10,000 data points are represented by 100 data points by coarse-graining.)
Figure 5
Residue positional fluctuations of the first 3 PCs in each dataset. Note that the PC 1 and PC 2 in the X-ray-I dataset have symmetrical fluctuations for the two protein chains (the first chain: residues 1–99; the second chain: residues 100–198). But no symmetrical fluctuations are observed for the X-ray-II, NMR and MD datasets.
Figure 6
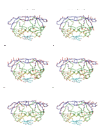
Visualizations of the motions of the dominant PCs (left column) and the most similar corresponding modes predicted by ENM (right column). In the X-ray-II dataset, the overlap between (A) PC 1 and (B) Mode 2 is 0.52. In the NMR dataset, the overlap between (C) PC 1 and (D) Mode 2 is 0.91. In the MD dataset, the overlap between (E) PC 1 and (F) Mode 1 is 0.74. Blue - the flap domain; green - the core domain; cyan - the terminal domain; yellow - other residues. The motions of PCs and modes are shown as red sticks with the directions indicated. The stick lengths represent the relative amplitudes of fluctuations of the corresponding residue.
Similar articles
- Distance matrix-based approach to protein structure prediction.
Kloczkowski A, Jernigan RL, Wu Z, Song G, Yang L, Kolinski A, Pokarowski P. Kloczkowski A, et al. J Struct Funct Genomics. 2009 Mar;10(1):67-81. doi: 10.1007/s10969-009-9062-2. Epub 2009 Feb 18. J Struct Funct Genomics. 2009. PMID: 19224393 Free PMC article. - The use of experimental structures to model protein dynamics.
Katebi AR, Sankar K, Jia K, Jernigan RL. Katebi AR, et al. Methods Mol Biol. 2015;1215:213-36. doi: 10.1007/978-1-4939-1465-4_10. Methods Mol Biol. 2015. PMID: 25330965 Free PMC article. - Probing structure-function relationships in human immunodeficiency virus type 1 protease via molecular dynamics simulation.
Harte WE Jr, Beveridge DL. Harte WE Jr, et al. Methods Enzymol. 1994;241:178-95. doi: 10.1016/0076-6879(94)41065-8. Methods Enzymol. 1994. PMID: 7854178 Review. - High-resolution modeling of protein structures based on flexible fitting of low-resolution structural data.
Zheng W, Tekpinar M. Zheng W, et al. Adv Protein Chem Struct Biol. 2014;96:267-84. doi: 10.1016/bs.apcsb.2014.06.004. Epub 2014 Aug 24. Adv Protein Chem Struct Biol. 2014. PMID: 25443961 Review.
Cited by
- Coarse grained normal mode analysis vs. refined Gaussian Network Model for protein residue-level structural fluctuations.
Park JK, Jernigan R, Wu Z. Park JK, et al. Bull Math Biol. 2013 Jan;75(1):124-60. doi: 10.1007/s11538-012-9797-y. Epub 2013 Jan 8. Bull Math Biol. 2013. PMID: 23296997 Free PMC article. - Structural compliance: A new metric for protein flexibility.
Scaramozzino D, Khade PM, Jernigan RL, Lacidogna G, Carpinteri A. Scaramozzino D, et al. Proteins. 2020 Nov;88(11):1482-1492. doi: 10.1002/prot.25968. Epub 2020 Jul 14. Proteins. 2020. PMID: 32548853 Free PMC article. - The landscape of the prion protein's structural response to mutation revealed by principal component analysis of multiple NMR ensembles.
Gendoo DM, Harrison PM. Gendoo DM, et al. PLoS Comput Biol. 2012;8(8):e1002646. doi: 10.1371/journal.pcbi.1002646. Epub 2012 Aug 9. PLoS Comput Biol. 2012. PMID: 22912570 Free PMC article. - Molecular determinants of cadherin ideal bond formation: Conformation-dependent unbinding on a multidimensional landscape.
Manibog K, Sankar K, Kim SA, Zhang Y, Jernigan RL, Sivasankar S. Manibog K, et al. Proc Natl Acad Sci U S A. 2016 Sep 27;113(39):E5711-20. doi: 10.1073/pnas.1604012113. Epub 2016 Sep 12. Proc Natl Acad Sci U S A. 2016. PMID: 27621473 Free PMC article. - Structural and Dynamic Characterizations Highlight the Deleterious Role of SULT1A1 R213H Polymorphism in Substrate Binding.
Dash R, Ali MC, Dash N, Azad MAK, Hosen SMZ, Hannan MA, Moon IS. Dash R, et al. Int J Mol Sci. 2019 Dec 11;20(24):6256. doi: 10.3390/ijms20246256. Int J Mol Sci. 2019. PMID: 31835852 Free PMC article.
References
- Amadei A, Ceruso MA, Di Nola A. On the convergence of the conformational coordinates basis set obtained by the essential dynamics analysis of proteins ’ molecular dynamics simulations. Proteins. 1999;36:419–424. - PubMed
- Amadei A, Linssen AB, Berendsen HJ. Essential dynamics of proteins. Proteins. 1993;17:412–425. - PubMed
- Amadei A, Linssen AB, de Groot BL, van Aalten DM, Berendsen HJ. An efficient method for sampling the essential subspace of proteins. J. Biomol. Struct. Dyn. 1996;13:615–625. - PubMed
- Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Folding Des. 1997;2:173–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM072014-03/GM/NIGMS NIH HHS/United States
- R01 GM072014/GM/NIGMS NIH HHS/United States
- GM-073095/GM/NIGMS NIH HHS/United States
- R01 GM073095/GM/NIGMS NIH HHS/United States
- GM-072014/GM/NIGMS NIH HHS/United States
- R01 GM072014-04/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources