Alternative splicing of Alu exons--two arms are better than one - PubMed (original) (raw)
Alternative splicing of Alu exons--two arms are better than one
Nurit Gal-Mark et al. Nucleic Acids Res. 2008 Apr.
Abstract
Alus, primate-specific retroelements, are the most abundant repetitive elements in the human genome. They are composed of two related but distinct monomers, left and right arms. Intronic Alu elements may acquire mutations that generate functional splice sites, a process called exonization. Most exonizations occur in right arms of antisense Alu elements, and are alternatively spliced. Here we show that without the left arm, exonization of the right arm shifts from alternative to constitutive splicing. This eliminates the evolutionary conserved isoform and may thus be selected against. We further show that insertion of the left arm downstream of a constitutively spliced non-Alu exon shifts splicing from constitutive to alternative. Although the two arms are highly similar, the left arm is characterized by weaker splicing signals and lower exonic splicing regulatory (ESR) densities. Mutations that improve these potential splice signals activate exonization and shift splicing from the right to the left arm. Collaboration between two or more putative splice signals renders the intronic left arm with a pseudo-exon function. Thus, the dimeric form of the Alu element fortuitously provides it with an evolutionary advantage, allowing enrichment of the primate transcriptome without compromising its original repertoire.
Figures
Figure 1.
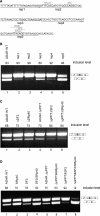
Characteristics of Alu right arm exons and their counterpart intronic left arms. (A) Alignment of the right and left arm of _Alu_J consensus sequence (gi551536) in its antisense orientation (relative to the mRNA) using the MAVID alignment server (73). The PPT of the right arm was extended to 19 nt as, on average, the PPT length in exonized _Alu_s is 19 bases ±3 (28) and is marked by horizontal brackets. The major 3′ss and 5′ss that are selected by Alu exons are indicated by arrows. Identical sequences are highlighted in gray. The 31-nt sequence that is present only in the right arm is indicated in a box. (B) Comparison of splice signals in the right and the left arms of 330 _Alu_s that have undergone exonization originating from the right arm in the antisense orientation. Boxes indicate right and left arms. Locations of the real and putative splice sites are marked by arrowheads. Distribution of ESEs, based on ESEfinder, throughout the exon and the putative splice sites in the left arm are shown by a histogram divided into four bins, each of which represents a quartile of the exon length. The number in the center of the boxes represents the mean ESE density score based on ESEfinder. (C) Comparison of ESR densities between the right and left Alu arms. ESR densities are shown for three ESR datasets: ESEfinder (38), Goren et al. (39) and Fairbrother et al. (40).
Figure 2.
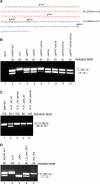
The intronic left arm of the Alu element is essential for the alternative splicing of an upstream exon. (A) The sequence of an Alu retroelement in its antisense orientation from which exon 8 of ADAR2 is exonized. The right arm (in red) undergoes exonization while the left arm (in blue) is intronic. The potential PPT, 3′ss, and 5′ss in the left arm (unexonized) are underlined. ESE densities (according to ESEfinder) for each arm are indicated on the right. (B) Plasmids containing the indicated mutants were transfected into 293T cells. Total cytoplasmic RNA was extracted, and splicing products were separated in 2% agarose gel after reverse transcription polymerase chain reaction (RT–PCR). Lane 1, vector only (pEGFP); lane 2, splicing products of wild-type ADAR2; and lanes 3–10, splicing products of ADAR2 minigene mutated at the left arm of the Alu element in different combinations. ΔLA: deletion of the entire intronic left arm; ΔpPPT: deletion of the 14-bp pPPT sequence of the left arm; Δp3′ss: elimination of two potential 3′ss in the intronic left arm (AGAG to ACCG); Δp5′ss: elimination of two potential 5′ss in the intronic left arm (GTGT to GAAT). The two possible minigene mRNA isoforms are shown on the right. The numbers above the lanes indicate exon 8 inclusion level. (C) Insertion of an intronic left arm of an Alu element downstream to a constitutive exon results in a shift toward alternative splicing. The sequence of the intronic left arm of the Alu element, from which exon 8 of ADAR2 gene undergoes exonization (in its antisense orientation), was cloned downstream to constitutive exon 12 in the IMP minigene. The splicing assay was performed as described above. Lane 1, splicing product of wild-type IMP; lanes 2–3, splicing products of IMP minigene in which the intronic left arm of the Alu element from ADAR2 was inserted downstream to exon 12 in antisense and sense orientations, respectively; lanes 4–5, splicing products of IMP minigene in which the intronic left arm of the Alu element from ADAR2 was inserted upstream to exon 12 in antisense and sense orientations, respectively. The two possible minigene mRNA isoforms are shown on the right. The numbers above the lanes indicate exon 12 inclusion levels. (D): Exonization of the intronic left arm. Splicing assays were performed as described above. Lane 1, splicing products of wild-type ADAR2; lane 2, splicing products of ADAR2 mutant harboring a deletion of the intronic left arm; lane 3, splicing products of ADAR2 mutant harboring a deletion of the exonized right arm; lane 4, splicing products of ADAR2 mutant after strengthening of the p5′ss and pPPT of the intronic left arm; lane 5, splicing products of ADAR2 mutant harboring a replacement of the intronic left arm with the sequence of an additional right arm. The two minigene mRNA isoforms are shown on the right and were confirmed by sequencing.
Figure 3.
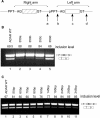
Increasing the distance between the left and right arms of the Alu element results in a shift toward constitutive splicing. (A) Illustration of the Alu element in its antisense orientation. The right and left arms are marked by horizontal brackets. An intronic sequence of 350 bp was inserted in four different positions along the intronic Alu's left arm: (a) between the exonized right arm and the intronic left arm; (b) between the pPPT and the p3′ss of the intronic left arm; (c) in the center of the intronic left arm; (d) downstream of the intronic left arm. (B) Splicing assays were performed as described in Figure 2. Lane 1, splicing products of wild-type ADAR2; lanes 2–5, splicing products of ADAR2 minigene mutated as indicated in A. (C) Intronic sequences of growing lengths (10–300 bp) were inserted between the exonized right arm and the intronic left arm. Lane 1, splicing products of wild-type ADAR2; lanes 2–11, splicing products of ADAR2 minigene mutated as indicated.
Figure 4.
The effect of pseudo splice signals and putative SR-binding sites within the left arm on the splicing pattern of the right arm. (A) Segments along the left arm 25 bp in length (rep1–rep5) were replaced with a designed sequence that did not contain any potential splice signals or SR-binding sites. The examined putative ESRs are shown. (B) Splicing assays were performed as described in Figure 2. Lane 2, splicing products of wild-type ADAR2; lanes 2–6, splicing products of ADAR2 minigene with replaced segments (rep1–rep5). (C) Lane 1, splicing products of wild-type ADAR2; lanes 2–6, splicing products of ADAR2 minigene mutated at the indicated binding sites in segments 2, with and without deletion of pPPT. (D) Changes in the binding capacities of putative SR proteins in segment 5 (near the potential 5′ss of the intronic left arm). Lane 1, splicing products of wild-type ADAR2; lanes 2–8, splicing products of ADAR2 minigene with the indicated mutations (see Supplementary Data).
Figure 5.
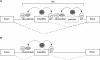
Why are Alu exons alternatively spliced? (A) The splicing pattern of exonization events originating from the right arm of Alu elements (ExonRA) is alternative due to the presence of a downstream arm that presumably acts as a pseudo-exon. Thus, competition between two highly similar arms shifts the splicing pattern of the stronger arm to alternative. Although the weaker left arm does not undergo exonization, it nonetheless participates in the regulation of the stronger right arm by reducing the affinity of the splicing machinery for the exonized arm, thereby causing it to be alternatively spliced. (B) However, when the exonized right arm (ExonRA) is free of left arm regulation it is constitutively spliced. Such events are presumably deleterious, and therefore may be selected against.
Similar articles
- Intronic Alus influence alternative splicing.
Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G. Lev-Maor G, et al. PLoS Genet. 2008 Sep 26;4(9):e1000204. doi: 10.1371/journal.pgen.1000204. PLoS Genet. 2008. PMID: 18818740 Free PMC article. - The birth of an alternatively spliced exon: 3' splice-site selection in Alu exons.
Lev-Maor G, Sorek R, Shomron N, Ast G. Lev-Maor G, et al. Science. 2003 May 23;300(5623):1288-91. doi: 10.1126/science.1082588. Science. 2003. PMID: 12764196 - Minimal conditions for exonization of intronic sequences: 5' splice site formation in alu exons.
Sorek R, Lev-Maor G, Reznik M, Dagan T, Belinky F, Graur D, Ast G. Sorek R, et al. Mol Cell. 2004 Apr 23;14(2):221-31. doi: 10.1016/s1097-2765(04)00181-9. Mol Cell. 2004. PMID: 15099521 - Exonization of transposed elements: A challenge and opportunity for evolution.
Schmitz J, Brosius J. Schmitz J, et al. Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review. - The origins and implications of Aluternative splicing.
Kreahling J, Graveley BR. Kreahling J, et al. Trends Genet. 2004 Jan;20(1):1-4. doi: 10.1016/j.tig.2003.11.001. Trends Genet. 2004. PMID: 14698612 Review.
Cited by
- Expressing genes do not forget their LINEs: transposable elements and gene expression.
Kines KJ, Belancio VP. Kines KJ, et al. Front Biosci (Landmark Ed). 2012 Jan 1;17(4):1329-44. doi: 10.2741/3990. Front Biosci (Landmark Ed). 2012. PMID: 22201807 Free PMC article. Review. - Intronic L1 retrotransposons and nested genes cause transcriptional interference by inducing intron retention, exonization and cryptic polyadenylation.
Kaer K, Branovets J, Hallikma A, Nigumann P, Speek M. Kaer K, et al. PLoS One. 2011;6(10):e26099. doi: 10.1371/journal.pone.0026099. Epub 2011 Oct 13. PLoS One. 2011. PMID: 22022525 Free PMC article. - The origins, evolution, and functional potential of alternative splicing in vertebrates.
Mudge JM, Frankish A, Fernandez-Banet J, Alioto T, Derrien T, Howald C, Reymond A, Guigó R, Hubbard T, Harrow J. Mudge JM, et al. Mol Biol Evol. 2011 Oct;28(10):2949-59. doi: 10.1093/molbev/msr127. Epub 2011 May 6. Mol Biol Evol. 2011. PMID: 21551269 Free PMC article. - Structural variants caused by Alu insertions are associated with risks for many human diseases.
Payer LM, Steranka JP, Yang WR, Kryatova M, Medabalimi S, Ardeljan D, Liu C, Boeke JD, Avramopoulos D, Burns KH. Payer LM, et al. Proc Natl Acad Sci U S A. 2017 May 16;114(20):E3984-E3992. doi: 10.1073/pnas.1704117114. Epub 2017 May 2. Proc Natl Acad Sci U S A. 2017. PMID: 28465436 Free PMC article. - Predicting Alu exonization in the human genome with a deep learning model.
He Z, Chen O, Phillips N, Pasquesi GIM, Sabunciyan S, Florea L. He Z, et al. bioRxiv [Preprint]. 2024 Jan 4:2024.01.03.574099. doi: 10.1101/2024.01.03.574099. bioRxiv. 2024. PMID: 38260329 Free PMC article. Preprint.
References
- Brett D, Pospisil H, Valcarcel J, Reich J, Bork P. Alternative splicing and genome complexity. Nat. Genet. 2002;30:29–30. - PubMed
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. - PubMed
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. - PubMed
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous