Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning - PubMed (original) (raw)
. 2008 Mar 13;452(7184):215-9.
doi: 10.1038/nature06745. Epub 2008 Feb 17.
Affiliations
- PMID: 18278030
- PMCID: PMC2377394
- DOI: 10.1038/nature06745
Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning
Shawn J Cokus et al. Nature. 2008.
Abstract
Cytosine DNA methylation is important in regulating gene expression and in silencing transposons and other repetitive sequences. Recent genomic studies in Arabidopsis thaliana have revealed that many endogenous genes are methylated either within their promoters or within their transcribed regions, and that gene methylation is highly correlated with transcription levels. However, plants have different types of methylation controlled by different genetic pathways, and detailed information on the methylation status of each cytosine in any given genome is lacking. To this end, we generated a map at single-base-pair resolution of methylated cytosines for Arabidopsis, by combining bisulphite treatment of genomic DNA with ultra-high-throughput sequencing using the Illumina 1G Genome Analyser and Solexa sequencing technology. This approach, termed BS-Seq, unlike previous microarray-based methods, allows one to sensitively measure cytosine methylation on a genome-wide scale within specific sequence contexts. Here we describe methylation on previously inaccessible components of the genome and analyse the DNA methylation sequence composition and distribution. We also describe the effect of various DNA methylation mutants on genome-wide methylation patterns, and demonstrate that our newly developed library construction and computational methods can be applied to large genomes such as that of mouse.
Figures
Figure 1. Methylation of different fractions of the Arabidopsis genome
a, Chromosome-wide distribution of methylation and correlation with repeats in sliding 100 kB windows. b, Methylation levels and siRNA abundance are plotted across different types of repeats and genes. c, High levels of methylation are detected at loci corresponding to siRNAs. d, Relationship between methylation levels and the length of different types of repeats and genes. e, From left to right, methylation levels of the three consecutive cytosines in the (CCCTAAA)n telomeric repeat unit are calculated in wild type and the drm1 drm2 cmt3 mutant, respectively.
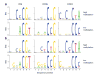
Figure 2. Sequence preferences for methylation in CG, CHG, and CHH contexts
Logos of sequence contexts that are preferentially methylated at the highest or lowest levels for 7-mer sequences in which the methylated cytosine is in the fifth position. In a, all genomic 7-mers in chromosome 1 were analyzed, while in b sequences were restricted to previously-defined methylated sequences . The logo graphically displays the sequence enrichment at a particular position in the alignment of 7-mers in each class, measured in bits. The maximum sequence conservation per site is 2 bits (i.e., 1 base) when a site is perfectly conserved, and 0 if there is no preference for a nucleotide.
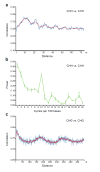
Figure 3. Methylation shows periodic patterns
a, c, Correlation of the methylation status of cytosines in a CHH (a) and CHG (c) context. The _x_-axis indicates the distance between the two cytosines. The _y_-axis indicates the level of autocorrelation in methylation. The red line is a running average of windows that are ±2 bases around a single base. b, Fourier transform analysis of CHH methylation correlation. The _x_-axis indicates the number of cycles per 100 bases. The _y_-axis is the amplitude of the corresponding frequency. The peak at position 10 represents a periodicity of ten nucleotides, with a _p_-value smaller than 10−108 for observing this periodicity value by chance in random permutations of the genome. In a-c, Monte Carlo sampling of three datasets each consisting of half the data was used to compute the mean and standard deviations of the autocorrelations and Fourier transforms. Mean values are shown and error bars (a and b) represent standard deviations. In a and b, methylation from the whole genome was analyzed, while in c the analysis was restricted to previously-defined methylated sequences (see Supplementary Fig. 15 for details).
Figure 4. BS-Seq profiling of methylation mutants in Arabidopsis and mouse
a, BS-Seq data mapping to protein-coding genes was plotted in 500 nucleotide sliding windows. Two vertical blue lines mark the boundaries between upstream regions and gene bodies (left) and between gene bodies and downstream regions (right). b, Distribution of methylation along chromosome 4 in 25 nucleotide sliding windows. In a and b, a horizontal blue line indicates zero percent methylation. c, Comparison of the amount of CG methylation in wild type and _mUhrf1_−/− embryonic stem cells, represented as the average number of CGs appearing per million sequenced nucleotides.
Similar articles
- Bisulphite sequencing of plant genomic DNA.
Aichinger E, Köhler C. Aichinger E, et al. Methods Mol Biol. 2010;655:433-43. doi: 10.1007/978-1-60761-765-5_29. Methods Mol Biol. 2010. PMID: 20734278 - Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis.
Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Zhang X, et al. Cell. 2006 Sep 22;126(6):1189-201. doi: 10.1016/j.cell.2006.08.003. Epub 2006 Aug 31. Cell. 2006. PMID: 16949657 - Analysis of DNA methylation in plants by bisulfite sequencing.
Foerster AM, Mittelsten Scheid O. Foerster AM, et al. Methods Mol Biol. 2010;631:1-11. doi: 10.1007/978-1-60761-646-7_1. Methods Mol Biol. 2010. PMID: 20204863 - Finding the fifth base: genome-wide sequencing of cytosine methylation.
Lister R, Ecker JR. Lister R, et al. Genome Res. 2009 Jun;19(6):959-66. doi: 10.1101/gr.083451.108. Epub 2009 Mar 9. Genome Res. 2009. PMID: 19273618 Free PMC article. Review. - DNA cytosine methylation in plant development.
Zhang M, Kimatu JN, Xu K, Liu B. Zhang M, et al. J Genet Genomics. 2010 Jan;37(1):1-12. doi: 10.1016/S1673-8527(09)60020-5. J Genet Genomics. 2010. PMID: 20171573 Review.
Cited by
- Challenges and Opportunities in Understanding Genetics of Fungal Diseases: Towards a Functional Genomics Approach.
Bruno M, Matzaraki V, van de Veerdonk FL, Kumar V, Netea MG. Bruno M, et al. Infect Immun. 2021 Jul 15;89(8):e0000521. doi: 10.1128/IAI.00005-21. Epub 2021 Jul 15. Infect Immun. 2021. PMID: 34031131 Free PMC article. Review. - Contiguous and stochastic CHH methylation patterns of plant DRM2 and CMT2 revealed by single-read methylome analysis.
Harris KD, Zemach A. Harris KD, et al. Genome Biol. 2020 Aug 6;21(1):194. doi: 10.1186/s13059-020-02099-9. Genome Biol. 2020. PMID: 32762764 Free PMC article. - An optimized algorithm for detecting and annotating regional differential methylation.
Li S, Garrett-Bakelman FE, Akalin A, Zumbo P, Levine R, To BL, Lewis ID, Brown AL, D'Andrea RJ, Melnick A, Mason CE. Li S, et al. BMC Bioinformatics. 2013;14 Suppl 5(Suppl 5):S10. doi: 10.1186/1471-2105-14-S5-S10. Epub 2013 Apr 10. BMC Bioinformatics. 2013. PMID: 23735126 Free PMC article. - Plants regenerated from tissue culture contain stable epigenome changes in rice.
Stroud H, Ding B, Simon SA, Feng S, Bellizzi M, Pellegrini M, Wang GL, Meyers BC, Jacobsen SE. Stroud H, et al. Elife. 2013 Mar 19;2:e00354. doi: 10.7554/eLife.00354. Elife. 2013. PMID: 23539454 Free PMC article. - Silicon era of carbon-based life: application of genomics and bioinformatics in crop stress research.
Li MW, Qi X, Ni M, Lam HM. Li MW, et al. Int J Mol Sci. 2013 May 29;14(6):11444-83. doi: 10.3390/ijms140611444. Int J Mol Sci. 2013. PMID: 23759993 Free PMC article. Review.
References
- Henderson IR, Jacobsen SE. Epigenetic Inheritance in Plants. Nature. 2007;447:418–424. - PubMed
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
- Zhang X, et al. Genome-wide High-Resolution Mapping and Functional Analysis of DNA Methylation in Arabidopsis. Cell. 2006;126:1189–201. - PubMed
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources