Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p - PubMed (original) (raw)
Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p
Zhiheng Yu et al. J Mol Biol. 2008.
Abstract
The type I AAA (ATPase associated with a variety of cellular activities) ATPase Vps4 and its co-factor Vta1p/LIP5 function in membrane remodeling events that accompany cytokinesis, multivesicular body biogenesis, and retrovirus budding, apparently by driving disassembly and recycling of membrane-associated ESCRT (endosomal sorting complex required for transport)-III complexes. Here, we present electron cryomicroscopy reconstructions of dodecameric yeast Vps4p complexes with and without their microtubule interacting and transport (MIT) N-terminal domains and Vta1p co-factors. The ATPase domains of Vps4p form a bowl-like structure composed of stacked hexameric rings. The two rings adopt dramatically different conformations, with the "upper" ring forming an open assembly that defines the sides of the bowl and the lower ring forming a closed assembly that forms the bottom of the bowl. The N-terminal MIT domains of the upper ring localize on the symmetry axis above the cavity of the bowl, and the binding of six extended Vta1p monomers causes additional density to appear both above and below the bowl. The structures suggest models in which Vps4p MIT and Vta1p domains engage ESCRT-III substrates above the bowl and help transfer them into the bowl to be pumped through the center of the dodecameric assembly.
Figures
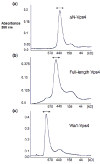
Fig. 1. Gel filtration analyses of multimeric Vps4p complexes
Multimeric ΔN-Vps4p, full-length Vps4p and Vta1p-Vps4p complexes were isolated by size – exclusion chromatography (Superose 6) in buffer A (100 mM NaCl, 25 mM Tris–HCl (pH 7.5), 2 mM MgCl2, 1 mM ATP, and 1mM DTT). Elution positions of molecular weight standards are shown below each chromatogram.
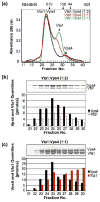
Fig. 2. The Vta1p-Vps4p complex exhibits 1:2 stoichiometry
(a) Size-exclusion chromatography of the complexes formed when Vta1p and Vps4p were mixed at molar ratios of 1:2 (black trace), 1:1 (red trace) and 3:1 (green trace). Samples were chromatographed on a Superdex 200 HR column in the presence of 1 mM ATP. For reference, elution positions of size standards and of free Vps4 and Vta1 proteins are shown above (arrows). (b) and (c) SDS-PAGE analyses (above) and associated protein quantities (below) for fractions spanning the Vps4p-Vta1p complex peaks shown in part (a) for the 1:2 and 1:1 mixtures, respectively.
Fig. 3. Cryo-EM images of full-length Vps4p
Before crosslinking, most regions of the cryo-grid had clear ice devoid of protein while other regions were filled with what appeared to be protein aggregates (top). After crosslinking (bottom), individual large protein complexes were routinely seen (example particles are circled). The hexagon encloses one complex with visible hexagonal symmetry. Scale bar 100 nm.
Fig. 4. Top views reveal 6-fold symmetry
Select class averages of (top) ΔN-Vps4p, (middle) full-length Vps4p, and (bottom) Vta1p-Vps4p complexes are shown. All the raw images were iteratively classified based on their similarity using singular value decomposition by refine2d.py. Particles within the same class were aligned and averaged to generate 50 class averages. The 18 class averages most closely corresponding to “top” views are shown, all of which suggest 6-fold symmetry.
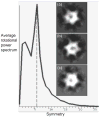
Fig. 5. Directed symmetry search
The EMAN program startcsym was used to search the particle gallery to find the images that were most apparently Cn-fold (n=5, 6, 7) symmetric. Three classes containing the 90 most 5- (panel a), 6- (panel b), and 7-fold (panel c) symmetric particles, respectively, were aligned and averaged. Even the most 5- and 7-fold symmetric particles appear 6-fold symmetric. The averaged rotational power spectrum of all 270 particles is also shown with a clear peak indicating 6-fold symmetry. Further statistical analysis by Rotastat also overwhelmingly favored 6-fold symmetry (not shown).
Fig. 6. 3-D cryo-EM reconstructions of ΔN-Vps4p, full-length Vps4p and Vta1p-Vps4p complexes
Isosurfaces are shown, contoured to enclose the full expected molecular weights of the ΔN-Vps4p, full-length Vps4p, and Vta1p-Vps4p complexes. All three complexes form bowl-like structures with a central cavity (asterisk) in the upper half, suggesting that the top and bottom rings are in very different conformations. The presence of the N-terminal MIT domain in full-length Vps4p causes a nipple (arrow) to appear above the cavity as well as six small fin-like densities (arrowheads) around the lower ring. The binding of Vta1p produces additional density mainly over and around the nipple and beneath the bottom ring. Scale bar 50 Å.
Fig. 7. Isosurface comparisons
(a) Full-length Vps4p contoured at 578 (solid purple) and 446 kDa (dashed purple) superimposed on ΔN-Vps4p contoured at 446 kDa (blue). The additional densities above the bowl and in the fins persist at both contours, indicating they are significant. (b) Vta1p-Vps4p complex contoured at 824 kDa (solid yellow line) and 578 kDa (innermost dashed yellow line) superimposed over the full-length Vps4p at 578 kDa (purple). Compared with the full-length Vps4p, the Vta1p-Vps4p complex has significant extra densities around and over the nipple above the top ring and beneath the center of the bottom ring. (c) and (d) Bottom and side view of the previously proposed model of hexameric Vps4B placed into the bottom ring of the full-length Vps4p reconstruction contoured at 578 kDa.
Fig. 8. Difference maps
(a) Full-length Vps4p minus ΔN-Vps4p, (b) Vta1p-Vps4p minus full-length Vps4p, and (c) Vta1p-Vps4p minus ΔN-Vps4p maps contoured at 3.5σ above the mean, superimposed on the 3-D reconstructions of the corresponding subcomplex. The largest difference densities are colored in purple (a) and yellow in (b) and (c). Additional smaller (shaded purple) or disconnected (gray) difference densities are also seen.
Fig. 9. Comparison of full-length Vps4p with cryo-EM reconstructions of type II AAA proteins
Full-length Vps4p, ClpB (EMDB ID 1243), NSF-αSNAP-SNARE (EMDB ID 1059), p97-p47 (EMDB ID 1191) and p97 (from E. M. Wilson-Kubalek) maps, all in presumably ATP-bound states, contoured at their full estimated molecular weights. The D2 rings of the type II AAA proteins face the viewer in the top row and comprise the lower rings in the bottom row.
Similar articles
- The oligomeric state of the active Vps4 AAA ATPase.
Monroe N, Han H, Gonciarz MD, Eckert DM, Karren MA, Whitby FG, Sundquist WI, Hill CP. Monroe N, et al. J Mol Biol. 2014 Feb 6;426(3):510-25. doi: 10.1016/j.jmb.2013.09.043. Epub 2013 Oct 23. J Mol Biol. 2014. PMID: 24161953 Free PMC article. - Vta1p and Vps46p regulate the membrane association and ATPase activity of Vps4p at the yeast multivesicular body.
Lottridge JM, Flannery AR, Vincelli JL, Stevens TH. Lottridge JM, et al. Proc Natl Acad Sci U S A. 2006 Apr 18;103(16):6202-7. doi: 10.1073/pnas.0601712103. Epub 2006 Apr 6. Proc Natl Acad Sci U S A. 2006. PMID: 16601096 Free PMC article. - Binding of Substrates to the Central Pore of the Vps4 ATPase Is Autoinhibited by the Microtubule Interacting and Trafficking (MIT) Domain and Activated by MIT Interacting Motifs (MIMs).
Han H, Monroe N, Votteler J, Shakya B, Sundquist WI, Hill CP. Han H, et al. J Biol Chem. 2015 May 22;290(21):13490-9. doi: 10.1074/jbc.M115.642355. Epub 2015 Apr 1. J Biol Chem. 2015. PMID: 25833946 Free PMC article. - Meiotic Clade AAA ATPases: Protein Polymer Disassembly Machines.
Monroe N, Hill CP. Monroe N, et al. J Mol Biol. 2016 May 8;428(9 Pt B):1897-911. doi: 10.1016/j.jmb.2015.11.004. Epub 2015 Nov 10. J Mol Biol. 2016. PMID: 26555750 Free PMC article. Review. - Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes.
McCullough J, Frost A, Sundquist WI. McCullough J, et al. Annu Rev Cell Dev Biol. 2018 Oct 6;34:85-109. doi: 10.1146/annurev-cellbio-100616-060600. Epub 2018 Aug 10. Annu Rev Cell Dev Biol. 2018. PMID: 30095293 Free PMC article. Review.
Cited by
- Biochemical and structural studies of yeast Vps4 oligomerization.
Gonciarz MD, Whitby FG, Eckert DM, Kieffer C, Heroux A, Sundquist WI, Hill CP. Gonciarz MD, et al. J Mol Biol. 2008 Dec 26;384(4):878-95. doi: 10.1016/j.jmb.2008.09.066. Epub 2008 Oct 4. J Mol Biol. 2008. PMID: 18929572 Free PMC article. - Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly.
Shim S, Merrill SA, Hanson PI. Shim S, et al. Mol Biol Cell. 2008 Jun;19(6):2661-72. doi: 10.1091/mbc.e07-12-1263. Epub 2008 Apr 2. Mol Biol Cell. 2008. PMID: 18385515 Free PMC article. - Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB.
Moriscot C, Gribaldo S, Jault JM, Krupovic M, Arnaud J, Jamin M, Schoehn G, Forterre P, Weissenhorn W, Renesto P. Moriscot C, et al. PLoS One. 2011;6(7):e21921. doi: 10.1371/journal.pone.0021921. Epub 2011 Jul 8. PLoS One. 2011. PMID: 21760923 Free PMC article. - Electron tomography of HIV-1 infection in gut-associated lymphoid tissue.
Ladinsky MS, Kieffer C, Olson G, Deruaz M, Vrbanac V, Tager AM, Kwon DS, Bjorkman PJ. Ladinsky MS, et al. PLoS Pathog. 2014 Jan 30;10(1):e1003899. doi: 10.1371/journal.ppat.1003899. eCollection 2014 Jan. PLoS Pathog. 2014. PMID: 24497830 Free PMC article. Clinical Trial. - The oligomeric state of the active Vps4 AAA ATPase.
Monroe N, Han H, Gonciarz MD, Eckert DM, Karren MA, Whitby FG, Sundquist WI, Hill CP. Monroe N, et al. J Mol Biol. 2014 Feb 6;426(3):510-25. doi: 10.1016/j.jmb.2013.09.043. Epub 2013 Oct 23. J Mol Biol. 2014. PMID: 24161953 Free PMC article.
References
- Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–12. - PubMed
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases