Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains - PubMed (original) (raw)
Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains
Hoanh Tran et al. Genes Dev. 2008.
Abstract
A key effector of the canonical Wnt pathway is beta-catenin, which binds to TCF/LEF factors to promote the transcription of Wnt target genes. In the absence of Wnt stimulation, beta-catenin is phosphorylated constitutively, and modified with K48-linked ubiquitin for subsequent proteasomal degradation. Here, we identify Trabid as a new positive regulator of Wnt signaling in mammalian and Drosophila cells. Trabid show a remarkable preference for binding to K63-linked ubiquitin chains with its three tandem NZF fingers (Npl4 zinc finger), and it cleaves these chains with its OTU (ovarian tumor) domain. These activities of Trabid are required for efficient TCF-mediated transcription in cells with high Wnt pathway activity, including colorectal cancer cell lines. We further show that Trabid can bind to and deubiquitylate the APC tumor suppressor protein, a negative regulator of Wnt-mediated transcription. Epistasis experiments indicate that Trabid acts below the stabilization of beta-catenin, and that it may affect the association or activity of the TCF-beta-catenin transcription complex. Our results indicate a role of K63-linked ubiquitin chains during Wnt-induced transcription.
Figures
Figure 1.
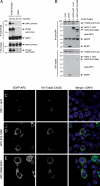
Association between Trabid and APC proteins. (A,B) Immunoprecipitations between Flag-dTrbd and Drosophila APC (HA-E-APC) (A) or Flag-Trabid and an N-terminal truncation of human APC, GFP-APC(1–1447) (B), in cotransfected 293 cells, as indicated in panels. (C_–_E) Immunofluorescence of 293 cells cotransfected with HA-Trabid C443S and different GFP-tagged APC fragments, as indicated, fixed and stained with α-HA antibody (red in merge) and DAPI (blue in merge, to reveal the nuclei; see also Supplemental Fig. S5). Recruitment of GFP-APC(1–1447) into C443S puncta was observed in 9% of transfected cells (n = 100); neither APC(918–1698) nor APC(2068–2843) colocalized with these puncta (n = 100).
Figure 2.
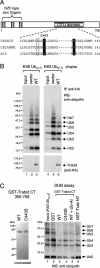
Cleavage of K63-linked ubiquitin by Trabid. (A) Domains of human Trabid and alignment of the invariant cysteines and histidines (shaded in black) of three human OTU family members, with putative catalytic cysteine (C443 in Trabid) and histidine residues shaded, and the D > A substitution characteristic of Trabid orthologs boxed. (B) DUB assays, with wild-type and mutant HA-tagged Trabid immunoprecipitated from transfected 293T cells, as indicated, incubated with K48- or K63-linked ubiquitin (Ub2–7). (C) In vitro DUB assays, with wild-type and mutant GST-tagged C- or N-terminal fragments of Trabid expressed in bacteria (left; see also Fig. 3C), incubated with K63-linked ubiquitin (Ub2–7; right). (Ub-Al) Ubiquitin aldehyde.
Figure 3.
Binding of Trabid’s NZF motifs to K63-linked ubiquitin. (A) Alignments of various NZF motifs, as indicated, with invariant cysteines shaded in black and other invariant residues in gray; the first cysteines (underlined) were mutated to alanine in the 3xCysNZF mutant. The T10 F/Y11 Φ22 signature for ubiquitin binding (Alam et al. 2004) is indicated by arrowheads (Φ, aliphatic amino acid; residue numbers refer to the position in this alignment); mutated TY residues are boxed. (B, Top) Ubiquitin-binding assays, with wild-type and mutant HA-Trabid immunoprecipitated from transfected 293T cells; asterisk indicates ubiquitylated protein coimmunoprecipitated with wild type and C443S; this signal is eliminated in the double-mutant C443S 3xNZFmut whose ubiquitin binding is abolished (see also D); background band, IgG heavy chain. (Below) The same blot was reprobed with α-HA antibody to monitor expression levels. (C) Ubiquitin-binding assays, with GST alone or GST-tagged Trabid(1–354) (see also Fig. 2C), expressed in bacteria (shown in bottom panel), and incubated in vitro with K48- or K63-linked ubiquitin; note the strong binding preference for K63-linked chains (shown in lanes 5,6). (D, top) Ubiquitin-binding assays as in C, with wild-type or mutant Trabid(1–200) (spanning the three tandem NZF motifs) incubated with K63-linked ubiquitin; TY > LV mutation of individual NZF (lanes 4_–_6), two NZF motifs (lanes 7_–_9), or all three NZF motifs (lane 10). (Middle panel) Long exposure of the same blot (bottom part only), to visualize residual binding of the NZF1 + 2 and NZF1 + 3 mutants to Ub4. (Bottom panel) The same blot was reprobed with α-GST antibody to monitor levels (revealing some breakdown; see also C). Note that the minimal module binding to K63-linked ubiquitin chains consists of at least two linked NZF motifs. (E,F) Immunofluorescence of 293 cells cotransfected with C443S and C443S-NZF1 + 2, fixed and stained as in Figure 1C–E.
Figure 4.
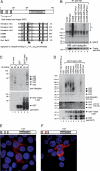
Requirement of Trabid for efficient TCF-mediated transcription. (A) Western blot showing siRNA depletion of endogenous Trabid in 293 cells (internal control, α-TLE). (B_–_D) Real-time quantitative RT–PCR assays, after transfection of 293 cells with siRNAs as in A, monitoring transcript levels of the Wnt target genes c-MYC, AXIN2, and BCL9, as indicated, after treatment of cells with control (L-CM) or Wnt3A-conditioned (W3a-CM) medium. (E) TOPFLASH luciferase assays, after two subsequent transfections of SW480 cells with siRNAs, with or without re-expression of wild-type and mutant HA-tagged ΔsiRNA Trabid rescue constructs, as indicated; control FOPFLASH values are also shown. Relative luciferase values are shown (statistical significance, [*] P < 0.01; [**] P < 0.05). (Below) Western blots showing HA-Trabid expression from one representative experiment (α-tubulin, loading controls). (F) NF-κΒ-dependent luciferase reporter assays in 293T cells transfected with siRNAs as in A and cotransfected with the expression vectors as indicated. (Below) Western blots showing expression levels. Error bars in this and subsequent figures indicate standard deviations from the mean, from two or three independent experiments (performed in duplicate).
Figure 5.
Epistasis analysis of Trabid’s function in the Wnt pathway. TOPFLASH assays in 293T cells transfected with siRNAs as in Figure 4F and cotransfected with empty vector, HA-Wnt3A (A), stabilized Flag-β-catenin (Δ45S) (B), or 50–100 ng of catC-LEF1Δ56 or 1–5 ng VP16-LEF1ΔN chimeras (C), as indicated (the expression levels of the two chimeras were chosen to result in comparable transactivation). (Below) Western blots from a representative experiment to monitor expression levels of endogenous or overexpressed proteins (α-β-catenin antibody was used to detect catC-LEF1Δ56; VP16-LEF1ΔN was undetectable at these low expression levels); numbers below the blot in C indicate relative levels of catC-LEF1Δ56 expression (normalized to lane 2).
Figure 6.
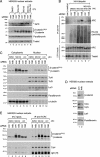
Hyperubiquitylation of APC in Trabid-depleted cells. Western blots of nuclear or cytoplasmic fractions 24 h after transfection of 293 (A,B,D) or 293T (C) cells transfected with siRNAs (A_–_D) (as in Fig. 4F) and HA-ubiquitin (B); treated for 4 h with DMSO (control), 10 mM MG132, and/or 20 mM LiCl or Wnt3A-conditioned medium as indicated; and probed sequentially with the antibodies indicated on the right. (E) Coimmunoprecipitations from nuclear fractions from 293T cells (200 μg), prepared as in C, as indicated.
Figure 7.
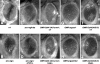
Function of dTrbd in the Wingless response of Drosophila. Eyes from control y w flies (A), or from flies after expression, in the larval eye disc of Wingless (B_–_D), Armadillo (E,F), Argos inhibitor of EGF receptor (G,H), or Delta agonist of Notch receptor (I,J). (A,B,E,G,I) +/+. (C,F,H,J) trbd/+. (D) dTCF3/+. trbd heterozygosity suppresses the rough eye phenotype due to ectopic Wingless or Armadillo (C,F), but not that due to ectopic Argos (H), Rhomboid (not shown), or Delta (J).
Similar articles
- Jerky/Earthbound facilitates cell-specific Wnt/Wingless signalling by modulating β-catenin-TCF activity.
Benchabane H, Xin N, Tian A, Hafler BP, Nguyen K, Ahmed A, Ahmed Y. Benchabane H, et al. EMBO J. 2011 Apr 20;30(8):1444-58. doi: 10.1038/emboj.2011.67. Epub 2011 Mar 11. EMBO J. 2011. PMID: 21399610 Free PMC article. - Chibby, a nuclear beta-catenin-associated antagonist of the Wnt/Wingless pathway.
Takemaru K, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. Takemaru K, et al. Nature. 2003 Apr 24;422(6934):905-9. doi: 10.1038/nature01570. Nature. 2003. PMID: 12712206 - Loss of Trabid, a new negative regulator of the drosophila immune-deficiency pathway at the level of TAK1, reduces life span.
Fernando MD, Kounatidis I, Ligoxygakis P. Fernando MD, et al. PLoS Genet. 2014 Feb 20;10(2):e1004117. doi: 10.1371/journal.pgen.1004117. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24586180 Free PMC article. - Wnt signaling inside the nucleus.
Shitashige M, Hirohashi S, Yamada T. Shitashige M, et al. Cancer Sci. 2008 Apr;99(4):631-7. doi: 10.1111/j.1349-7006.2007.00716.x. Epub 2008 Jan 2. Cancer Sci. 2008. PMID: 18177486 Free PMC article. Review. - Signaling through beta-catenin and Lef/Tcf.
Novak A, Dedhar S. Novak A, et al. Cell Mol Life Sci. 1999 Oct 30;56(5-6):523-37. doi: 10.1007/s000180050449. Cell Mol Life Sci. 1999. PMID: 11212302 Free PMC article. Review.
Cited by
- Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme.
Ovaska K, Laakso M, Haapa-Paananen S, Louhimo R, Chen P, Aittomäki V, Valo E, Núñez-Fontarnau J, Rantanen V, Karinen S, Nousiainen K, Lahesmaa-Korpinen AM, Miettinen M, Saarinen L, Kohonen P, Wu J, Westermarck J, Hautaniemi S. Ovaska K, et al. Genome Med. 2010 Sep 7;2(9):65. doi: 10.1186/gm186. Genome Med. 2010. PMID: 20822536 Free PMC article. - The deubiquitinase ZRANB1 is an E3 ubiquitin ligase for SLC7A11 and regulates ferroptotic resistance.
Huang S, Zhang Q, Zhao M, Wang X, Zhang Y, Gan B, Zhang P. Huang S, et al. J Cell Biol. 2023 Nov 6;222(11):e202212072. doi: 10.1083/jcb.202212072. Epub 2023 Oct 13. J Cell Biol. 2023. PMID: 37831441 Free PMC article. - Evolutionary Loss of Activity in De-Ubiquitylating Enzymes of the OTU Family.
Louis M, Hofmann K, Broemer M. Louis M, et al. PLoS One. 2015 Nov 20;10(11):e0143227. doi: 10.1371/journal.pone.0143227. eCollection 2015. PLoS One. 2015. PMID: 26588485 Free PMC article. - Deubiquitinating Enzyme-Mediated Signaling Networks in Cancer Stem Cells.
Kaushal K, Ramakrishna S. Kaushal K, et al. Cancers (Basel). 2020 Nov 4;12(11):3253. doi: 10.3390/cancers12113253. Cancers (Basel). 2020. PMID: 33158118 Free PMC article. Review. - Chemical ubiquitination for decrypting a cellular code.
Stanley M, Virdee S. Stanley M, et al. Biochem J. 2016 May 15;473(10):1297-314. doi: 10.1042/BJ20151195. Biochem J. 2016. PMID: 27208213 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases